TOP
 Research
Research
 Research Teams for Science of Computing and Science by Computing
Research Teams for Science of Computing and Science by Computing
 Computational Structural Biology Research Team
Computational Structural Biology Research Team
Computational Structural Biology Research Team
Japanese
Team Principal Florence Tama
 florence.tama[at]riken.jp (Lab location: Kobe)
florence.tama[at]riken.jp (Lab location: Kobe)- Please change [at] to @
- 2015
- Professor, Graduate School of Science, Nagoya University(-present)
- 2013
- team Leader, Computational Structural Biology Research team, AICS (renamed R-CCS in 2018), RIKEN(-present)
- 2006
- Assistant Professor, University of Arizona, USA
- 2001
- Post-doctoral researcher, The Scripps Research Institute, USA
- 2000
- Ph.D. in Computational Biophysics, University Paul Sabatier, France
Keyword
- Modeling
- Structure
- Function
- Dynamics
- Low-resolution
Research summary
Biological complexes, structured ensembles of proteins or proteins/nucleic acids, perform many vital cellular functions, such as gene transcription, protein synthesis or regulation of cellular transport, and dysfunctions of those result in severe diseases. In order to understand diseases and develop treatments, the functional mechanisms of these biological complexes need to be elucidated. A crucial step in this process is the characterization of the structure and dynamics of these complexes. As large complexes are difficult to study by X-ray crystallography, alternative low-resolution experimental techniques such as cryo-electron microscopy, small angle X-ray scattering and fluorescence resonance energy transfer are often used to characterize their conformational states. Additionally, new experimental developments of X-ray Free Electron Lasers (XFEL, such as in SPring 8, RIKEN) should provide structural information close to atomic resolution. Our goal is to develop computational methods to obtain atomic level description of the functional states of biological complexes. Such methods will rely on the integration of various experimental data such as high resolution X-ray crystallography, lower resolution cryo-EM and emerging near atomic resolution XFEL with computational modeling through high performance computing such as with the supercomputers K and Fugaku. In addition, we aim to collaborate with experimental groups and the pharmaceutical industry to elucidate functional mechanism of biological systems.
Main research results
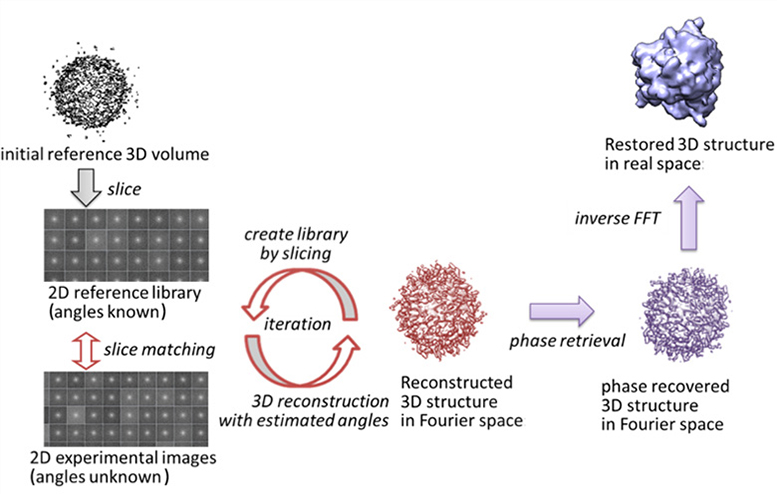
Restoring 3D structures from XFEL diffraction data
Recent development of intense XFEL light sources offers the possibility to obtain new structural information of biological macromolecules, in particular imaging of single macromolecules. Atomic level resolution such as obtained by X-ray crystallography cannot yet be achieved given current experimental conditions. But as XFEL experiments are still undergoing development for routine applications, computational algorithms to understand and analyze experimental data also need to be developed. In particular, to restore the 3D structures of biomolecules from the 2D diffraction patterns obtained by XFEL experiments, computational algorithms are necessary to estimate the laser beam incidence-angles to the molecule.
We are working on a program package for XFEL analysis that is based on XMIPP, commonly used software for image processing of single-particle 3D cryo-electron microscopy. Since XMIPP is designed to work with 2D data in real space, some of the routines were modified to handle 2D data in Fourier space. Through an iterative procedure, the orientations of the biomolecules that each image represents are estimated, and the 3D structure factor-amplitude is reconstructed. The 3D model in real space is obtained by phase retrieval. This approach was successfully tested with experimental data similar to XFEL diffraction patterns taken from a single nanoparticle.
Representative papers
- T. Nagai, F. Tama & O. Miyashita.:
"Cryo-cooling effect on DHFR crystal studied by replica-exchange molecular dynamics simulations"
Biophysical J. 116(3):395-405 (2019). - T. Mori, M. Kulik, O. Miyashita, J. Jung, F. Tama and Y. Sugita.:
"Acceleration of cryo-EM flexible fitting by efficient space partitioning for large biomolecular systems"
Structure 27(1):161-174. (2019). - A. Srivastava, F. Tama, D. Kohda & O. Miyashita.:
"Computational Investigation of the Conformational Dynamics in Tom20-Mitochondrial Presequence Tethered Complexes"
Proteins 87(1):81-90. (2019). - T. Nagai, Y. Mochizuki, Y. Joti, F. Tama & O. Miyashita.:
"Gaussian mixture model for coarse-grained modeling from XFEL"
Optics Express 26(20):26734-26749. (2018). - S. P. Tiwari, F. Tama & O. Miyashita.:
"Searching for 3D structural models from a library of biological shapes using a few 2D experimental images"
BMC Bioinformatics 19:320. (2018). - M. Nakano, O. Miyashita, S. Jonic, A. Tokuhisa, F. Tama.:
"Single-particle XFEL 3D Reconstruction of Ribosome-size Particles based on Fourier Slice Matching: Requirements to Reach Subnanometer Resolution"
J. Synchrotron Rad. 25:1010-1021. (2018). - O. Miyashita, C. Kobayashi, T. Mori, Y. Sugita, F. Tama.:
"Flexible Fitting to Cryo-EM Density Map using Ensemble Molecular Dynamics Simulations"
J. Comp. Chem. 38:1447-1461. (2017). - A. Tokuhisa, S. Jonic, F. Tama and O. Miyashita.:
"Hybrid Approach for Structural Modeling of Biological Systems from X-ray Free Electron Laser Diffraction Patterns"
J. Struct. Biol. 194(3):325-336 (2016). - M. Gallagher-Jones, Y. Bessho, S. Kim, J. Park, S. Kim, D. Nam, C. Kim, Y. Kim, Y. Noh do, O. Miyashita, F. Tama, Y. Joti, T. Kameshima, T. Hatsui, K. Tono, Y. Kohmura, M. Yabashi, SS. Hasnain, T. Ishikawa, C. Song.:
"Macromolecular structures probed by combining single-shot free-electron laser diffraction with synchrotron coherent X-ray imaging"
Nature Commun. 2:5:3798. (2014). -
Q Jin, CO Sorzano, JM de la Rosa-Trevín, JR Bilbao-Castro, R. Núñez-Ramírez, O. Llorca , F. Tama, S. Jonić.:
"Iterative elastic 3D-to-2D alignment method using normal modes for studying structural dynamics of large macromolecular complexes"
Structure 22:496-506. (2014).
