Tutorials of REMD simulations
Because complex biomolecules have many local-minimum energy states, conventional MD simulations often get trapped in one of the local minima, resulting in that unreliable free energy profile is obtained. With GENESIS program, we can perform several types of REMD simulations. Among them, we illustrate how to perform temperature REMD and REUS simulations and how to analyze the trajectories of the simulations here.
2.1 Temperature REMD simulation of a small peptide
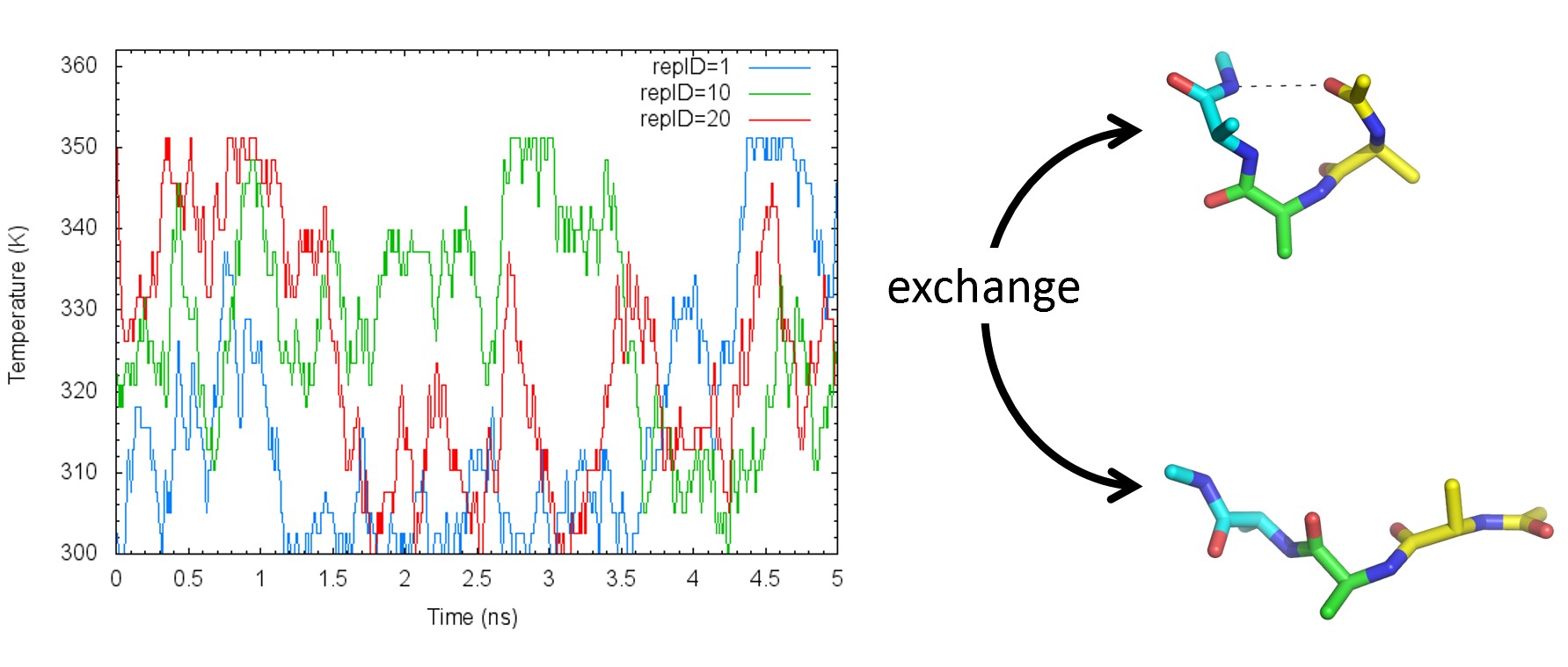
Temperature replica-exchange molecular dynamics (T-REMD) simulation method is used to achieve good sampling of systems with rugged free energy surfaces by increasing and decreasing the tempreratures of replicas. Here, we perform a T-REMD simulation of the alanine tripeptide in water. We also demonstrate how to analyze the trajectory generated with T-REMD simulations.
2.2 Replica-exchange umbrella sampling
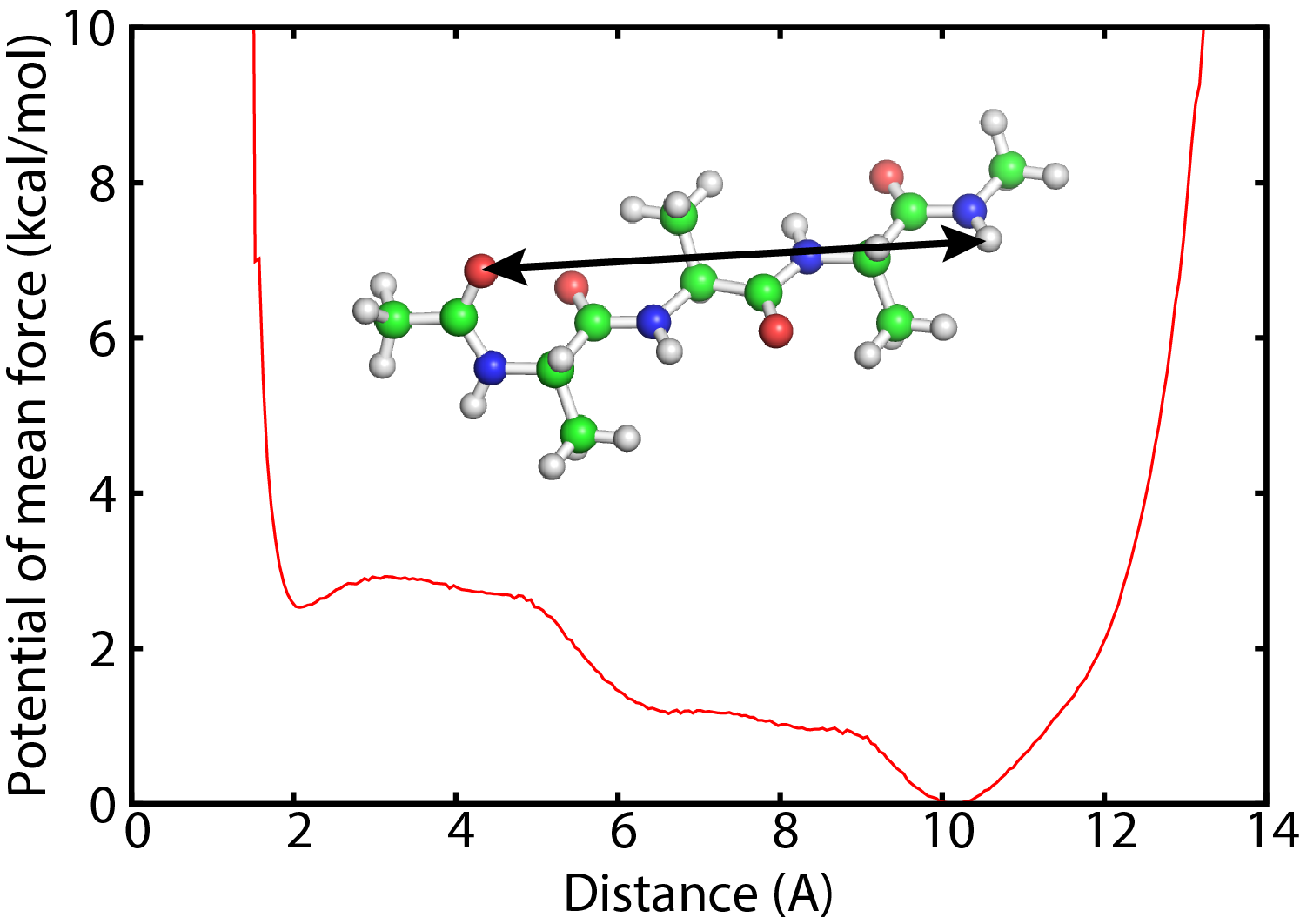
Umbrella sampling method has been widely used to calculate free energy profile. In this tutorial, we demonstrate a replica-exchange umbrella sampling simulation (REUS) of the alanine tripeptide in water. We analyze the free energy profile of the end-to-end distance of the peptide.