Tutorials of basic MD simulations
1.1 All-atom MD simulation of BPTI in NaCl solution
In this tutorial, we introduce MD simulations of a small protein BPTI in NaCl solution with the CHARMM force field. We explain how to build the initial structure and how to carry out energy minimization, equilibration, and production run by using GENESIS.
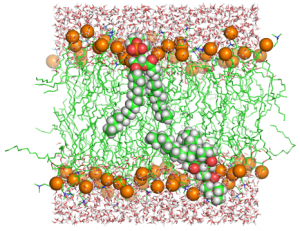
1.2 All-atom MD simulation of POPC lipid bilayers
Cell membranes have many biological roles such as substrate transport, signal transduction, and cell adhesion. By using GENESIS, the user can simulate biological membranes at the atomistic level. One of the difficulties in such simulations is initial setup of the bilayer structure or equilibration of the system. In this tutorial, we explain how to simulate POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine) lipid bilayers with GENESIS and CHARMM-GUI.
1.3 Coarse-grained MD simulation with the Go model
Coarse-graining makes long-time and large-scale simulations possible, which would be difficult with all-atom MD. This tutorial shows how to construct a coarse-grained representation of a protein, run coarse-grained simulation in GENESIS, and conduct simple analysis.