Removing ring penetrations and chirality errors
Contents
In this page, we explain how to remove ring penetrations and chirality errors in a protein using GENESIS. Here, we use spdyn. The same scheme is available in atdyn. The scheme in the below figure will be performed [1]. For details, please read the GENESIS user manual ([MINIMIZE] chapter).
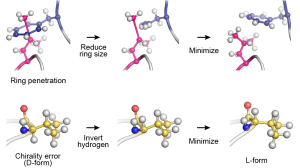
Preparation
Let’s download the tutorial file (tutorial22-A.6.zip). This tutorial is mainly composed of two steps: 1) system setup and 2) energy minimization. The energy minimization is further composed of three steps to remove ring penetrations and chirality errors. Control files for GENESIS are already included in the download file. Since we use the CHARMM36m force field parameters, we make a symbolic link to the CHARMM toppar directory (see Tutorial 2.2).
# Put the tutorial file in the Works directory $ cd ~/GENESIS_Tutorials-2022/Works $ mv ~/Downloads/tutorial22-A.6.zip ./ $ unzip tutorial22-A.6.zip # Let's clean up the directory $ mv tutorial22-A.6.zip TRASH # Let's take a note $ echo "tutorial-A.6: Remove ring penetration and chirality errors" >> README # Check the contents in Appendix 6 $ cd tutorial-A.6 $ ln -s ../../Data/Parameters/toppar_c36_jul21 ./toppar $ ln -s ../../Programs/genesis-2.0.0/bin ./bin $ ls INP1 INP2 INP3 bin setup toppar
# Setup the initial PDB and PSF files
$ cd setup
$ ls
build.tcl proa.pdb
$ vmd -e build.tcl
$ ls
build.tcl ionized.psf protein.pdb wbox.log wbox.psf
ionized.pdb proa.pdb protein.psf wbox.pdb
$ cd ../
Energy minimization
Step1: Run basic minimization
First, we carry out a basic energy minimization for this system.
# Check the control file
$ less INP1
[MINIMIZE]
method = SD # [SD]
nsteps = 5000 # number of minimization steps
crdout_period = 1000
rstout_period = 5000
# Run minimization
$ export OMP_NUM_THREADS=4
$ mpirun -np 8 ./bin/spdyn INP1 > log1
$ ls
INP1 INP2 INP3 log1 min1.dcd min1.rst bin setup toppar
$ less log1
However, in the log message you can see that there are several residues that may have ring penetrations or chirality errors in the energy minimized structure.
Check_Ring_Structure> Check ring structure
suspicious ring group id = 12 : TRP 68 (atom = 1011) max_bond_length = 1.921
WARNING!
Some suspicious residues were detected. Minimization might be too short, or "ring penetration"
might happen in the above residues. Check the structure of those residues very carefully.
If you found a ring penetration, try to perform an energy minimization again
with the options "check_structure = YES" and "fix_ring_error = YES" in [MINIMIZE].
The energy minimization should be restarted from the restart file obtained in "this" run.
For more information, see the chapter on [MINIMIZE] in the user manual.
Check_Chirality> Check chirality
suspicious chiral group id = 5 : GLU 5 (atom = 62) angle = 178.549
suspicious chiral group id = 32 : PRO 32 (atom = 488) angle = 85.871
WARNING!
Some suspicious residues were detected. Minimization might be too short, or "chirality error"
might happen in the above residues. Check the structure of those residues very carefully.
If you found a chirality error, try to perform an energy minimization again
with the options "check_structure = YES" and "fix_chirality_error = YES" in [MINIMIZE].
The energy minimization should be restarted from the restart file obtained in "this" run.
For more information, see the chapter on [MINIMIZE] in the user manual.
Let’s check the corresponding residues carefully using a molecular viewer software.

Step2: Run minimization to remove errors
In order to remove the ring penetrations and chirality errors, we restart the energy minimization using min1.rst. Let’s take a look at INP2. In the [MINIMIZE] section, fix_ring_error and fix_chirality_error options are added. Basically, these options should be added only when the error was found in the energy minimized structure.
[INPUT]
topfile = ./toppar/top_all36_prot.rtf
parfile = ./toppar/par_all36m_prot.prm
strfile = ./toppar/toppar_water_ions.str
psffile = ./setup/ionized.psf
pdbfile = ./setup/ionized.pdb
rstfile = min1.rst
[MINIMIZE]
method = SD # [SD]
nsteps = 5000 # number of minimization steps
crdout_period = 1000
rstout_period = 5000
fix_ring_error = YES
fix_chirality_error = YES
Run the energy minimization again using INP2.
# Run minimization
$ mpirun -np 8 ./bin/spdyn INP2 > log2
$ less log2
However, unfortunately, we could not fix the chirality error in Pro32 in this run.
Check_Ring_Structure> Check ring structure
No suspicious residue was detected.
Check_Chirality> Check chirality
suspicious chiral group id = 32 : PRO 32 (atom = 488) angle = 171.247
WARNING!
Some suspicious residues were detected. Minimization might be too short, or "chirality error"
might happen in the above residues. Check the structure of those residues very carefully.
If you found a chirality error, try to perform an energy minimization again
with the options "check_structure = YES" and "fix_chirality_error = YES" in [MINIMIZE].
The energy minimization should be restarted from the restart file obtained in "this" run.
For more information, see the chapter on [MINIMIZE] in the user manual.
Step3: Second trial to remove the remaining errors
So, we run the energy minimization again using INP3, in which fix_chirality_error = YES is specified.
[INPUT]
topfile = ./toppar/top_all36_prot.rtf
parfile = ./toppar/par_all36m_prot.prm
strfile = ./toppar/toppar_water_ions.str
psffile = ./setup/ionized.psf
pdbfile = ./setup/ionized.pdb
rstfile = min2.rst
[MINIMIZE]
method = SD # [SD]
nsteps = 5000 # number of minimization steps
crdout_period = 1000
rstout_period = 5000
fix_chirality_error = YES
# Run minimization
$ mpirun -np 8 ./bin/spdyn INP3 > log3
$ less log3
Now, you can see that there are no suspicious residues. Please take a look at Trp68, Glu5, and Pro32 to check whether the errors were actually removed. Then, you can use the obtained restart file (min3.rst) for the subsequent MD simulation.
Check_Ring_Structure> Check ring structure
No suspicious residue was detected.
Check_Chirality> Check chirality
No suspicious residue was detected.
References
- T. Mori et al., J. Chem. Inf. Model., 61, 3516–3528 (2021).
Written by Takaharu Mori@RIKEN Theoretical molecular science laboratory
June 16, 2022
