4.4 How to make a simulation movie with PyMol?
Contents
Computational biophysicists often show simulation movies in their oral presentations at seminars, conferences, and so on. In order to show a movie in a presentation, you need to create a movie file from MD trajectories and embed it in your presentation slides. In this tutorial, we will explain how to output images of molecular structures in MD trajectories using PyMol, one of the famous molecular viewers in the field of structural biology, and how to create a movie file by combining these images. We will use the convert and ffmpeg commands as well as GENESIS. The convert command is available in the Imagemagick software package. These tools should be installed before this tutorial.
Outline
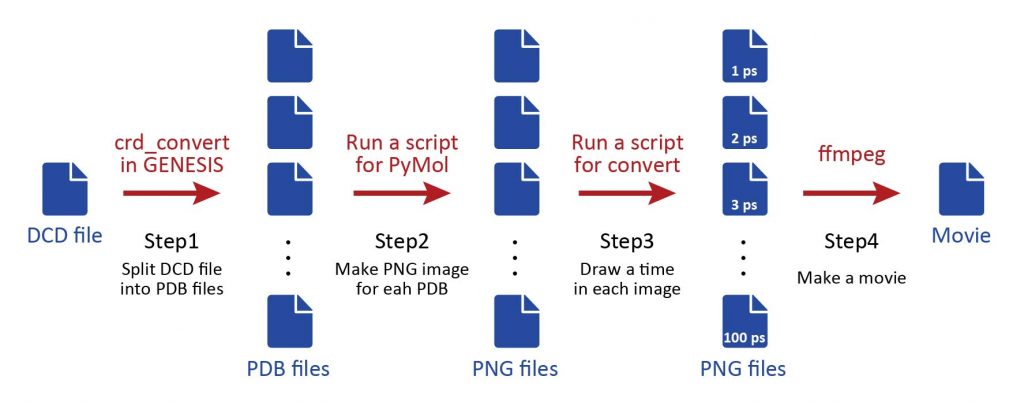
The scheme consists of four steps shown in the figure below: In Step1, the DCD file obtained from the MD simulation is split into individual PDB files using the GENESIS crd_convert tool. In Step2, PyMol is run with a script to create png images of each snapshot. In Step3, run the convert command again with a script to write the simulation time to each image. Finally, in Step4, use the ffmpeg command to combine all the images and create a move file.
Preparation
Let’s download the PDB and DCD sample files (tutorial22-4.4.zip) for this tutorial. This is the MD trajectory of a small protein, and the trajectory contains 100 frames. The sample does not contain any water molecules, but you can use the same protocol even if solvent is explicitly present. The input and script files for each step are already included in the respective directories.
# Download the tutorial file
$ cd ~/GENESIS_Tutorials-2022/Works
$ mv ~/Downloads/tutorial22-4.4.zip ./
$ unzip tutorial22-4.4.zip
# Let's clean up the directory
$ mv tutorial22-4.4.zip ./TRASH
# Let's take a note
$ echo "tutorial-4.4: Movie making using PyMol" >> README
$ cd tutorial-4.4
$ ln -s ../../Programs/genesis-2.0.0/bin ./
1_makepdb 2_makepng 3_convert 4_ffmpeg Data bin
# Check the trajectory data and initial structure
$ ls ./Data/
md.dcd proa.pdb
Step1. Create individual PDB files from the DCD file
First, we will split the DCD file into individual PDB files using the GENESIS crd_convert tool. Before we run the tool, let’s take a look at the control file.
# Change the directory for Step1
$ cd 1_makepdb
# Take a look at the control file of crd_convert
$ less INP
[OUTPUT] pdbfile = md0.pdb trjfile = md{}.pdb [OPTION] trjout_format = PDB # (PDB/DCD) trjout_type = COOR+BOX # (COOR/COOR+BOX) trjout_atom = 2 # atom group split_trjpdb = YES # output split PDB trajectory
The most important options in the control file are shown above and colored in red: “split_trjpdb = YES” and “trjout_format = PDB” in the [OPTION] section, and “trjfile = md{}.pdb” in the [OUTPUT] section. The index of the snapshot will be inserted in parentheses in the output file name. Let’s execute the crd_convert tool to this control file.
# Run the crd_convert tool
$ ../bin/crd_convert INP > log
$ ls
INP md2.pdb md33.pdb md47.pdb md60.pdb md74.pdb md88.pdb
log md20.pdb md34.pdb md48.pdb md61.pdb md75.pdb md89.pdb
md0.pdb md21.pdb md35.pdb md49.pdb md62.pdb md76.pdb md9.pdb
md1.pdb md22.pdb md36.pdb md5.pdb md63.pdb md77.pdb md90.pdb
md10.pdb md23.pdb md37.pdb md50.pdb md64.pdb md78.pdb md91.pdb
:
Step2. Create an image file for each snapshot
In Step2, we will execute PyMol to all the PDB files generated in Step1. In the Step2 directory, there are three files input_0.pml, template.pml, and Make.csh. First, let’s take a look at input_0.pml. This is a short PyMol script, which just reads ../1_makepdb/md0.pdb. Execute PyMol using this script.
# Change the directory for Step2 $ cd ../2_makepng $ ls Make.csh input_0.pml template.pml # Take a look at the PyMol script for md0.pdb $ less input_0.pml # Run PyMol $ pymol input_0.pml
Rotate the molecule in the PyMol window to determine the orientation of the molecule you like. Then, press the “Get View” button in the upper right corner, and you will see that matrix-like values are output in the terminal or in the upper window. These values are actually corresponding to the rotation matrix, position of the camera, and so on. We will use these values later.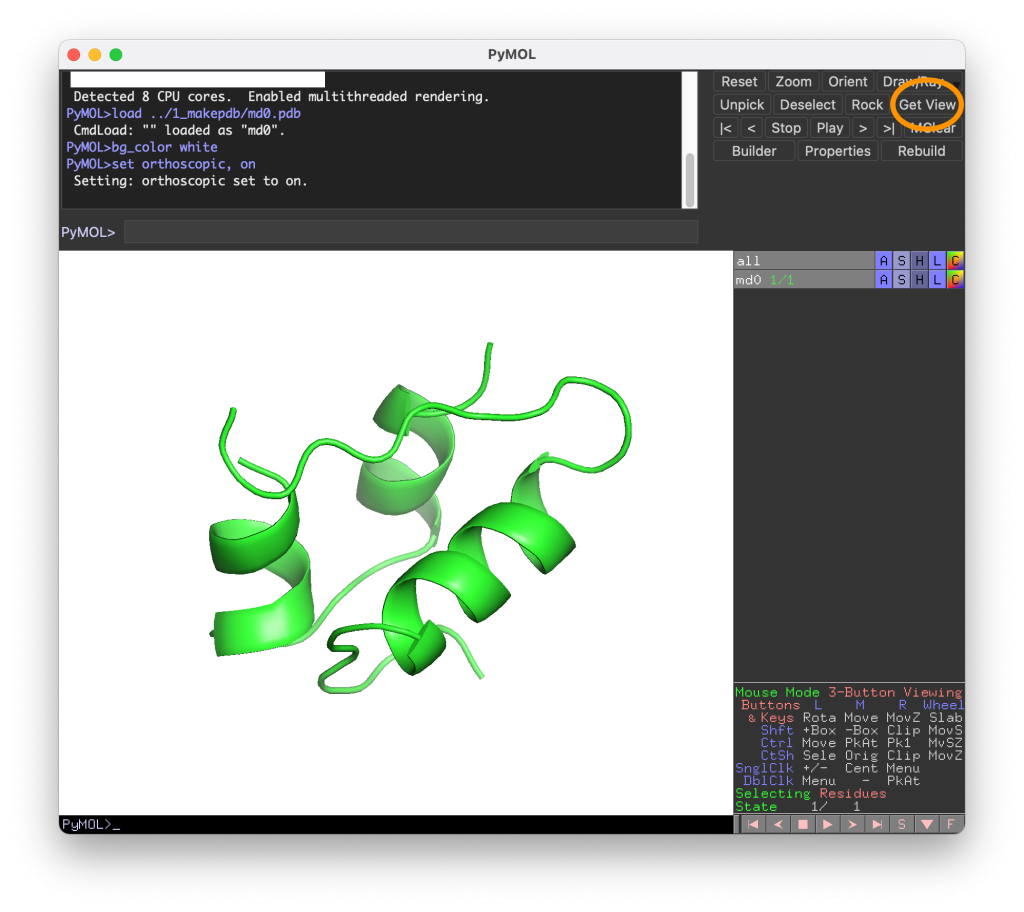
set_view (\
0.700811803, 0.321628720, -0.636726499,\
-0.506851256, 0.852599680, -0.127194136,\
0.501961410, 0.411864907, 0.760529041,\
-0.000017758, -0.000041828, -106.357391357,\
-18.747209549, 0.224934518, -8.418970108,\
86.687660217, 126.027915955, 20.000000000 )
Next, let’s take a look at template.pml. By using this file as a template, we will make a new PyMol script file (input_all.pml; see below) that will output an image for all PDB files. In template.pml, you will find the strings AAA and BBB, which will be replaced later by Make.csh with the indexes of the PDB file. In the middle part of the file, we write the set_view (...), which is corresponding to the above values. The width and height of a PNG image are specified by the variables width and height, respectively. Feel free to edit template.pml for your favorite representation by yourselves (see PyMol manual).
# Take a look at the script $ less template.pml
# Set view width = 480 height = 480 cmd.viewport(width, height) bg_color white set ray_shadow, off set ray_opaque_background, on set_view (\ 0.700811803, 0.321628720, -0.636726499,\ -0.506851256, 0.852599680, -0.127194136,\ 0.501961410, 0.411864907, 0.760529041,\ -0.000017758, -0.000041828, -106.357391357,\ -18.747209549, 0.224934518, -8.418970108,\ 86.687660217, 126.027915955, 20.000000000 )
Next, let’s take a look at Make.csh. Here, the variables ista and iend represent the first and last indexes of the PDB file where we create the image. You can also see that there is a “while” statement, which is an iterative loop that replaces the strings AAA and BBB in template.pml with the PDB indexes, and writes them to a single file input_all.pml.
# Take a look at the script for making input_all.pml
$ less Make.csh
#!/bin/csh set ista = 0 set iend = 100 echo -n > input_all.pml @ i = $ista while ($i <= $iend) set IDX = `echo "$i" | awk '{printf("%04d",$1)}'` sed -e "s/AAA/$i/g" template.pml | sed -e "s/BBB/$IDX/g" >> input_all.pml @ i = $i + 1 end
Let’s run Make.csh and take a look at the generated input_all.pml. You can see that template.pml is actually copied as a template in input_all.pml, where the commands to load md0.pdb to md100.pdb and output their images are written. Finally, let’s execute PyMol for input_all.pml with the -c option (command line mode). You will get the image files md0000.png through md0100.png.
# Run the script $ ./Make.csh $ ls Make.csh input_0.pml input_all.pml template.pml # Check the PyMol script $ less input_all.pml # Run PyMol $ pymol -c input_all.pml $ ls Make.csh md0015.png md0033.png md0051.png md0069.png md0087.png input_0.pml md0016.png md0034.png md0052.png md0070.png md0088.png input_all.pml md0017.png md0035.png md0053.png md0071.png md0089.png md0000.png md0018.png md0036.png md0054.png md0072.png md0090.png md0001.png md0019.png md0037.png md0055.png md0073.png md0091.png :
md0100.png
Step3. Write a simulation time in each image
In Step 3, we will write the simulation time in the image. Displaying the time of each snapshot helps the viewer to understand the time line of the simulation video. Here, we will use a script again to handle a large number of image files. If you take a look at Run.csh in the Step3 directory, you can see that it executes the convert command on all the PNG files.
# Change directory for Step3
$ cd ../3_convert
$ ls
Run.csh
# Take a look at the script
$ less Run.csh
#!/bin/csh
set ista = 0
set iend = 100
@ i = $ista
while ($i <= $iend)
set IDX = `echo "$i" | awk '{printf("%04d",$1)}'`
# Calculate the simulation time of each snapshot
set simtim = `echo "$i" | awk '{printf("%3d",$1)}'`
echo "IDX = $IDX simtim = $simtim"
# run the convert command
convert -font Nimbus-Mono-Bold -pointsize 32 \
-gravity northeast -fill "black" \
-draw "text 30,20 '${simtim} ps'" \
../2_makepng/md$IDX.png md$IDX.png
@ i = $i + 1
end
The options of the convert command determine what characters are drawn in the image and how they are drawn. Here, we draw the simulation time ($simtim) in the upper right corner (-gravity northeast). Since monospaced fonts are convenient for time display, we select “Nimbus-Mono-Bold” (to see the available fonts, type “convert -list font” on the command line). In this example, the time is the same as the “snapshot index”. In other cases, you will need to calculate the correct simtim in the awk command (e.g., awk '{printf("%5.1f",$1*1.5)}', if the time interval is 1.5 ps) or something similar.
# Run the script
$ ./Run.csh
$ ls
Run.csh md0016.png md0033.png md0050.png md0067.png md0084.png
md0000.png md0017.png md0034.png md0051.png md0068.png md0085.png
md0001.png md0018.png md0035.png md0052.png md0069.png md0086.png
md0002.png md0019.png md0036.png md0053.png md0070.png md0087.png
md0003.png md0020.png md0037.png md0054.png md0071.png md0088.png
:
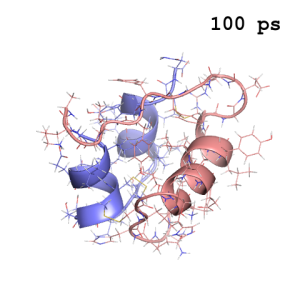
md0100.png
Step4. Combine image files into a movie file
Finally, we run the ffmpeg command on the all images obtained in Step3 to create a movie file. The following is an example of creating a movie in AVI format and converting it to MP4. The resulting movie is shown below.
# Change directory for Step4
$ cd ../4_ffmpeg
# Run the ffmpeg command
$ ffmpeg -i ../3_convert/md%4d.png -qscale 0 -vcodec mjpeg -s 480x480 md.avi
# Convert the file format from avi to mp4 (if needed)
$ ffmpeg -i md.avi -b 4000k -vcodec libx264 -pix_fmt yuv420p md.mp4
Written by Takaharu Mori@RIKEN Theoretical molecular science laboratory
Feb 25, 2022
