10.1. Acceleration of MD simulations using HMR and a longer time step
Contents
Here we show an example how to apply a longer time step than usual by making use of the hydrogen mass repartitioning (HMR) scheme with group temperature/pressure. Please note that this option is only available in GENESIS v2.0.
1. Overview of Hydrogen mass repartitioning (HMR)
In hydrogen mass repartitioning (HMR), the mass of hydrogen atoms is scaled by 2~3 while the mass of the heavy atom bonded to the hydrogen is reduced such that the total mass is not changed. The figure below is an example of HMR with a scaling ratio = 3. Here, numbers in each atom mean atomic mass.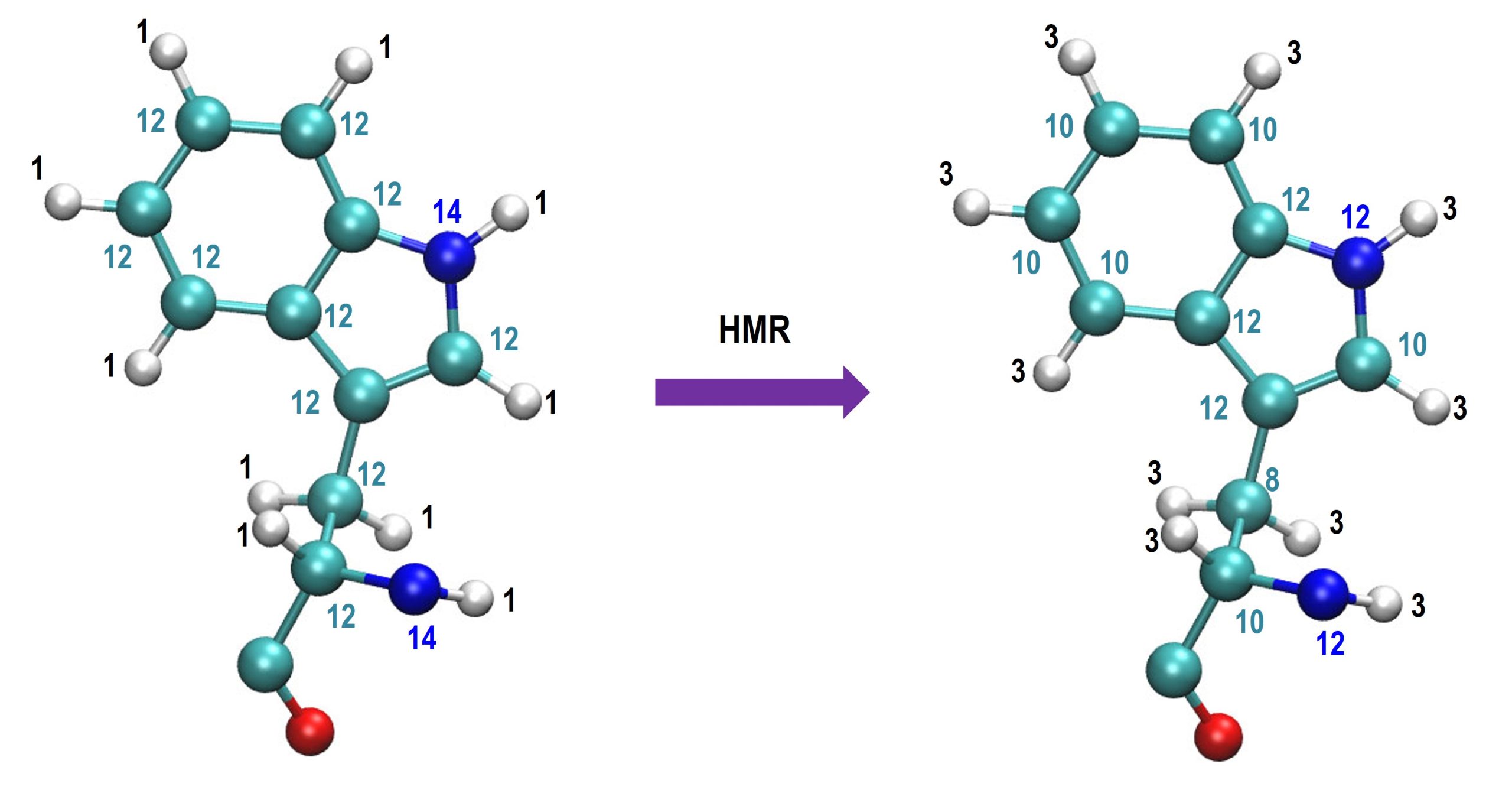
According to our recent investigation, the HMR scaling of hydrogen atoms in five- or six-member rings should be 2 to increase the stability with a large time step, as shown in the figure below:
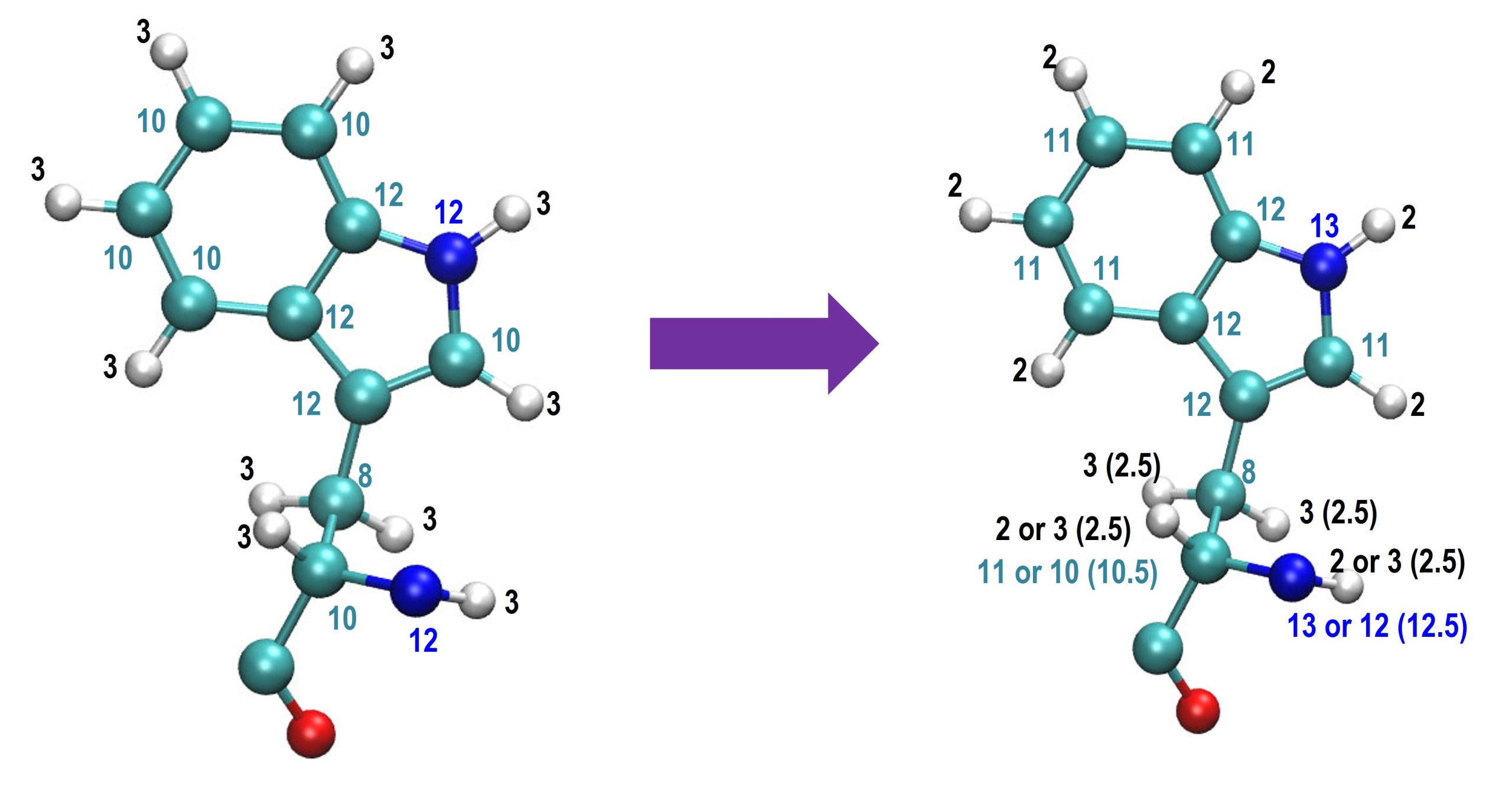
For other hydrogen atoms, we recommend using a HMR scaling ratio of 3 and 2.5 for CHARMM and AMBER force fields, respectively. In addition, it is better not to apply HMR to water molecules if you are going to observe dynamic properties from MD. In this tutorial, we will show how to make use of HMR in the simulation of CI2 protein solvated in water molecules. All the input files used in this tutorial can be downloaded in tutorial22-10.1.zip.
2. Preparations
2.1. Download/Compile GENESIS source code (v2.0beta)
GENESIS v2.0beta source code can be downloaded from github. After downloading it, you can compile with the following procedure.
$ cd genesis
$ autoreconf
$ ./configure --enable-single
$ make
$ make install
In the case of mixed precision, you can write –-enable-mixed instead of –-enable-single. For double precision, you don’t need to add anything. If you are interested in accurate integration with good energy conservation, we recommend to compile with mixed or double precisions.
Open MPI or Intel MPI libraries can be chosen by writing
# Open MPI
$ ./configure --enable-single FC=mpif90 CC=mpicc
# Intel MPI
$ ./configure --enable-single FC=mpiifort CC=mpiicc
To compile GENESIS on Fugaku or GPU machines, please write as followings:
# Fugaku
$ ./configure --enable-single --host=Fugaku
# GPU
$ ./configure --enable-single –with-cuda={your path of CUDA library}
2.2. Download Tutorial
The input files in this tutorial cover from minimization to production run. After downloading the tutorial file (tutorial22-10.1.zip), you can see five directories and two files.
$ unzip tutorial22-10.1.zip
$ cd tutorial-10.1
$ ls
1.min 3.equil_hmr step3_pbcsetup.pdb toppar
2.equil 4.production step3_pbcsetup.psf
3. MD simulation of CI2
To perform the minimization and equilibration runs, you can just follow Level2: STANDARD MD Tutorials. Here, we will present just a brief explanation.
3.1. Minimization
Minimization is the first required step to avoid instability in the structure. The input file is in the 1.min directory.
$ cd 1.min
$ ls
inp
The content of the Input file is
[INPUT]
parfile = ../toppar/par_all36m_prot.prm
strfile = ../toppar/toppar_all36_prot_c36m_d_aminoacids.str, ../toppar/toppar_water_ions.str
psffile = ../step3_pbcsetup.psf
pdbfile = ../step3_pbcsetup.pdb
reffile = ../step3_pbcsetup.pdb
[OUTPUT]
rstfile = min.rst
[ENERGY]
forcefield = CHARMM
electrostatic = PME
switchdist = 10.0
cutoffdist = 12.0
pairlistdist = 13.5
pme_ngrid_x = 64
pme_ngrid_y = 64
pme_ngrid_z = 64
pme_nspline = 4
water_model = NONE
vdw_force_switch = YES
contact_check = YES
nonbond_kernel = generic
[MINIMIZE]
method = SD
nsteps = 1000
rstout_period = 1000
[BOUNDARY]
type = PBC
box_size_x = 70
box_size_y = 70
box_size_z = 70
[SELECTION]
group1 = (sid:PROA) and backbone
group2 = (sid:PROA) and not backbone and not hydrogen
[RESTRAINTS]
nfunctions = 2
function1 = POSI
constant1 = 1
select_index1 = 1
function2 = POSI
constant2 = 0.1
select_index2 = 2
In minimization, the contact_check option is often used to avoid errors when we use an unstable structure in input. In GENESIS v2.0, the nonbond_kernel is automatically decided from the hardware. However when contact_check option is used, the generic kernel, which is identical to the one used in GENESIS 1.7, is assigned.
3.2. Equilibration
In this tutorial, we run equilibration with two steps after the minimization. First, let’s move to the 2.equil directory.
$ cd ../2.equil
$ ls
inp1 inp2
Here, there are two input files. In inp1, we run the equilibration with a small time step and with positional restraints. Therefore, there is no difference between inp1 and minimization input in [INPUT], [SELECTION], [RESTRAINTS], and [BOUNDARY]. The other sections in inp1 are written in the following way:
[OUTPUT]
rstfile = equil1.rst
dcdfile = equil1.dcd
[ENERGY]
forcefield = CHARMM
electrostatic = PME
switchdist = 10.0
cutoffdist = 12.0
pairlistdist = 13.5
pme_ngrid_x = 64
pme_ngrid_y = 64
pme_ngrid_z = 64
pme_nspline = 4
vdw_force_switch = YES
[DYNAMICS]
integrator = VVER
timestep = 0.001
nsteps = 100000
crdout_period = 5000
eneout_period = 1000
rstout_period = 100000
nbupdate_period = 10
[CONSTRAINTS]
rigid_bond = YES
[ENSEMBLE]
ensemble = NPT
tpcontrol = BUSSI
temperature = 303.15
pressure = 1.0
In the [ENERGY] section, the contact_check option is not written based on the assumption that the unstable structure disappeared after the minimization. rigid_bond in the [CONSTRAINTS] section is defined to be YES to avoid the vibrational motion of hydrogen atoms. After running with inp1, you can continue the equilibration with inp2 in which positional restraints are removed and with 2 fs time step is assigned.
3.3. Equilibration with HMR
If we run an MD simulation with HMR, you can see that the temperature value at the initial step is deviated from the final step of the previous equilibration. To avoid unstable result at initial steps with HMR, it is first recommended to run equilibration with HMR or not to include production results for the first a few ns results. In this tutorial, we will follow the first approach. To make use of HMR, let’s move to 3.equil_hmr directory.
$ cd ../3.equil_hmr/
$ ls
inp
The input file, inp, is similar to inp2 in 2.equil directory except
hydrogen_mr = yes
hmr_ratio = 3.0
hmr_ratio_xh1 = 2.0
hmr_target = solute
Here, we scale the mass of hydrogen atoms in the following manner: three times for for XH2 or XH3 and twice for XH1 (where X and H represent any heavy and hydrogen atoms, repsectively). In addition, HMR is not applied water molecules. To make use of the HMR option, first please write hydrogen_mr = yes. If not, HMR is not applied even if you write HMR related keys in the control input file. The HMR scaling ratio is controlled by hmr_ratio. It scales up the mass of hydrogen atoms while the mass of the bonded heavy atom is reduced to conserve the total mass. hmr_ratio_xh1 scales the mass of hydrogen atoms in XH1.
Please note that HMR input in GENESIS is distinguished from NAMD or AMBER program. In NAMD and AMBER, the scaled hydrogen mass should be written directly in the psf and prmtop files, respectively. For example, the psf files with and without HMR scaling are written as following:
#psf without HMR
32413 !NATOM
1 PROA 20 MET N NH3 -0.300000 14.0070 0 0.00000 -0.301140E-02
2 PROA 20 MET HT1 HC 0.330000 1.00800 0 0.00000 -0.301140E-02
3 PROA 20 MET HT2 HC 0.330000 1.00800 0 0.00000 -0.301140E-02
4 PROA 20 MET HT3 HC 0.330000 1.00800 0 0.00000 -0.301140E-02
5 PROA 20 MET CA CT1 0.210000 12.0110 0 0.00000 -0.301140E-02
6 PROA 20 MET HA HB1 0.100000 1.00800 0 0.00000 -0.301140E-02
. . .
psf with HMR
32413 !NATOM
1 PROA 20 MET N NH3 -0.300000 7.95900 0 0.00000 -0.301140E-02
2 PROA 20 MET HT1 HC 0.330000 3.02400 0 0.00000 -0.301140E-02
3 PROA 20 MET HT2 HC 0.330000 3.02400 0 0.00000 -0.301140E-02
4 PROA 20 MET HT3 HC 0.330000 3.02400 0 0.00000 -0.301140E-02
5 PROA 20 MET CA CT1 0.210000 9.99500 0 0.00000 -0.301140E-02
6 PROA 20 MET HA HB1 0.100000 3.02400 0 0.00000 -0.301140E-02
. . .
In this example, the masses of hydrogen atoms with HMR become three times larger (1.008 to 3.024) in the psf file, while the masses of boned heavy atoms are reduced accordingly. Therefore, psf files should be regenerated to perform MD with HMR. Similarly, masses in an AMBER parameter file are changed to apply HMR:
# parameter without HMR
%FLAG MASS
%FORMAT(5E16.8)
1.40070000E+01 1.00800000E+00 1.00800000E+00 1.00800000E+00 1.20110000E+01
1.00800000E+00 1.20110000E+01 1.00800000E+00 1.00800000E+00 1.20110000E+01
. . .
# parameter with HMR
%FLAG MASS
%FORMAT(5E16.8)
7.95900000E+00 3.02400000E+00 3.02400000E+00 3.02400000E+00 9.99500000E+00
3.02400000E+00 7.97900000E+00 3.02400000E+00 3.02400000E+00 7.97900000E+00
Unlike them, we do not have to change psf or parameter files. Instead we just write the HMR option, and program automatically performs simulations with scaled hydrogen masses.
3.4. Production run with HMR
After you equilibrate with HMR, you can run MD simulations with a large time step. Let’s move to the directory of production runs (4.production).
$ cd ../4.production/
$ ls
inp1 inp2 inp2_nohmr
In inp1, we perform production run with multiple time step with 3.5 fs for the short time step and 7.0 fs for the long time step. In inp2 and inp2_nohmr, we perform production run with 5 fs time step with and without HMR, respectively. Main differences in the control inputs are shown as following:
inp1:
[DYNAMICS]
integrator = VRES
timestep = 0.0035
nsteps = 120000
crdout_period = 3000
eneout_period = 60
rstout_period = 120000
nbupdate_period = 6
hydrogen_mr = yes
hmr_ratio = 3.0
hmr_ratio_xh1 = 2.0
hmr_target = solute
thermostat_period = 6
barostat_period = 6
inp2:
[DYNAMICS]
integrator = VVER
timestep = 0.005
nsteps = 100000
crdout_period = 2000
eneout_period = 50
rstout_period = 100000
nbupdate_period = 4
hydrogen_mr = yes
hmr_ratio = 3.0
hmr_ratio_xh1 = 2.0
hmr_target = solute
thermostat_period = 4
barostat_period = 4
inp2_nohmr:
[DYNAMICS]
integrator = VVER
timestep = 0.005
nsteps = 100000
crdout_period = 2000
eneout_period = 50
rstout_period = 100000
nbupdate_period = 4
thermostat_period = 4
barostat_period = 4
Here, we note that crdout_period (period of trajectory writing output), rstout_period (period of restart file writing output), nbupdate_period (period of pairlist updating), thermostat_period (period of thermostat), and barostat_period (period of barostat) should be adjusted such that the total update interval should not be changed.
The effect of HMR can be understood by running inp2 and inp2_nohmr. We could run inp2 without any problem. On the other hand, running inp2_nohmr has an error message in constraints:
Compute_Shake> SHAKE algorithm failed to converge: indexes 775 776 777
In other words, the MD simulation with a large time step is not stable without HMR assignment.
With optimal temperature estimation, atomic temperature and pressure evaluations require iterations when rigid_bod=yes is used in [CONSTRAINTS]. To avoid the iteration, we recommend using group temperature and pressure evaluations by writing group_tp=yes in the [ENSEMBLE] section.
Reference
- Jaewoon Jung, Kento Kasahara, Chigusa Kobayashi, Hiraku Oshima, Takaharu Mori, Yuji Sugita, “Optimized hydrogen mass repartition scheme combined with accurate temperature/pressure evaluations for thermodynamic and kinetic properties of biological systems”, J. Chem. Theory Comput. 17, 5312-5321 (2021) [Link]
- Jaewoon Jung, Yusji Sugita, “Group-based evaluation of temperature and pressure for molecular dynamics simulation with a large time step”, J. Chem. Phys. 153, 234115 (2020) [Link]
- Jaewoon Jung, Chigusa Kobayashi, Yuji Sugita, “Optimal temperature evaluation in molecular dynamics simulations with a large time step”, J. Chem. Theory Copmput. 15, 84-94 (2019) [Link]
- Jaewoon Jung, Chigusa Kobayashi, Yuji Sugita, “Kinetic energy definition in velocity Verlet integration for accurate pressure evaluation”, J. Chem. Phys. 148, 164109 (2018) [Link]
Written by Jaewoon Jung@RIKEN R-CCS. March, 2022
Updated by Jaewoon Jung@RIKEN R-CCS. June, 23, 2022
