Sample control file
Two-dimensional REMD (T-REMD/REUS) simulation
Two-dimensional REMD (T-REMD/REUS) simulation can be done by just combining Temperature exchange with REUS or Restraint exchange with T-REMD in the [REMD] section. The following sample achieves the T-REMD/REUS simulation, where temperature is exchanged in the range of 300.00-332.99 K with 8 replicas and also distance restraint is exchanged with 6 replicas (8×6=48 replicas in total).
[INPUT]
topfile = top_all36_prot.rtf # topology file
parfile = par_all36_prot.prm # parameter file
strfile = toppar_water_ions.str # stream file
psffile = ionized.psf # protein structure file
pdbfile = ionized.pdb # input PDB file
rstfile = eq_rep{}.rst # restart file
[OUTPUT]
logfile = run_rep{}.log # log file
dcdfile = run_rep{}.dcd # coordinates trajectory file
rstfile = run_rep{}.rst # restart file
remfile = run_rep{}.rem # parameter index file
[REMD]
dimension = 2 # Number of dimension of REMD simulation
exchange_period = 2500 # attempt exchange every 5 ps
type1 = TEMPERATURE # Temperature exchange
nreplica1 = 8 # Number of replicas
parameters1 = 300.00 304.54 309.14 313.80 318.51 323.28 328.10 332.99
type2 = RESTRAINT # restraint function is exchanged
nreplica2 = 6 # number of replicas
rest_function2 = 1 # target restraint function index
[ENERGY]
forcefield = CHARMM # CHARMM force field
electrostatic = PME # Particle Mesh Ewald method
switchdist = 10.0 # switch distance
cutoffdist = 12.0 # cutoff distance
pairlistdist = 13.5 # pair-list distance
vdw_force_switch = YES # force switch option for van der Waals
pme_nspline = 4 # order of B-spline in [PME]
pme_max_spacing = 1.0 # max grid spacing
[DYNAMICS]
integrator = LEAP # Leapfrog Verlet integrator
nsteps = 50000 # number of MD steps
timestep = 0.002 # timestep (ps)
eneout_period = 100 # energy output period
crdout_period = 100 # coordinates output period
rstout_period = 50000 # restart output period
nbupdate_period = 10 # nonbond update period
[CONSTRAINTS]
rigid_bond = YES # constraints all bonds involving hydrogen
[ENSEMBLE]
ensemble = NVT # NVT ensemble
tpcontrol = LANGEVIN # thermostat
gamma_t = 1.0 # thermostat friction coefficient (ps-1)
[BOUNDARY]
type = PBC # periodic boundary condition
[SELECTION]
group1 = an:N and resno: 1 # select atom i
group2 = an:O and resno: 10 # select atom j
[RESTRAINTS]
nfunctions = 1
function1 = DIST # use distance restraint
select_index1 = 1 2 # restrained atom index i and j
constant1 = 10 10 10 10 10 10 # force constant of each replica
reference1 = 4.0 4.3 4.5 4.7 4.9 5.1 # target distance of each replica
In the T-REMD/REUS simulation with this control file, the following exchange scheme is used:
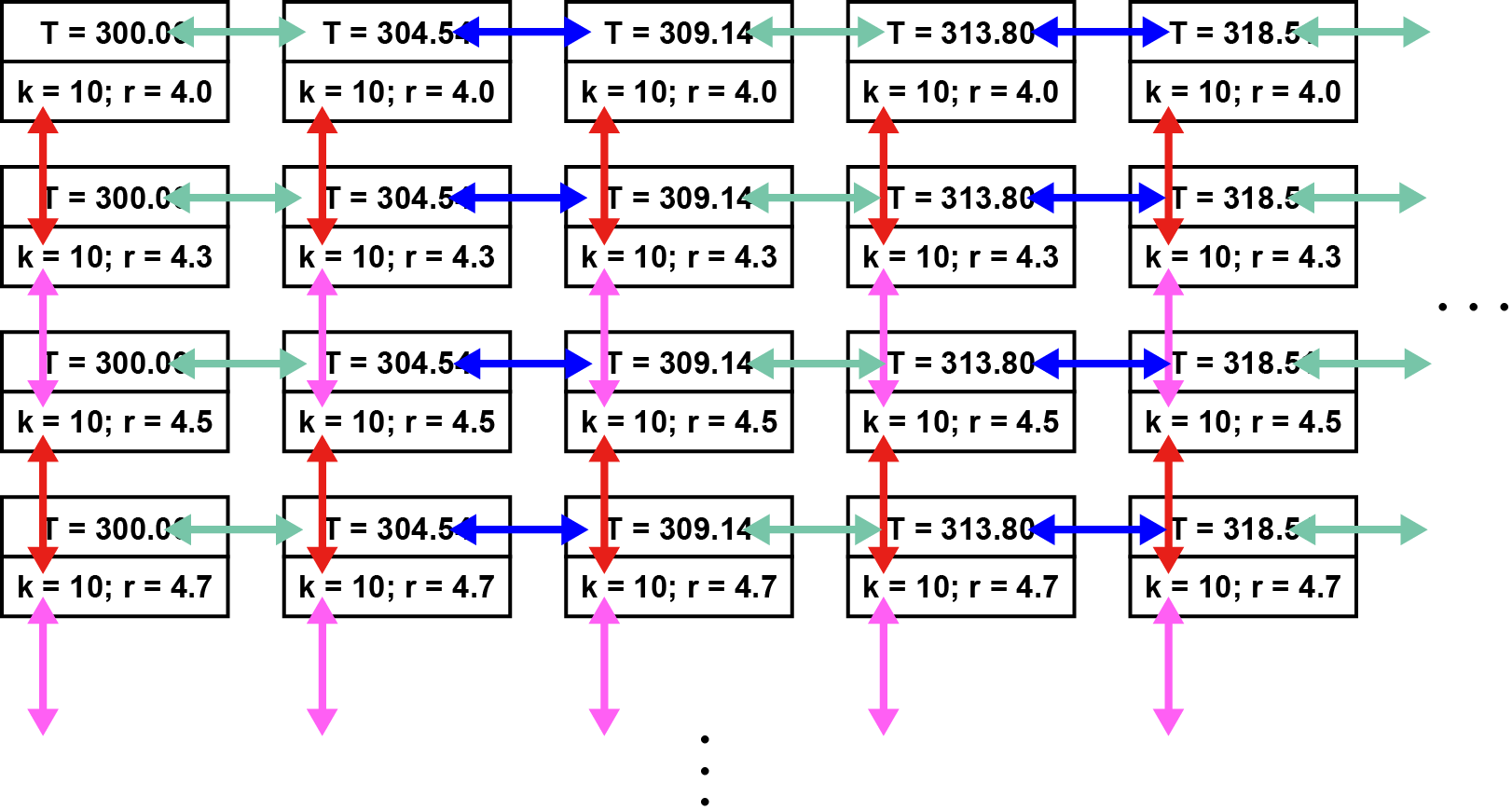
2500 step: 2nd dimension among (1↔2), (3↔4), … pairs (Restraint exchange between red pairs)
5000 step: 1st dimension among (2↔3), (4↔5), … pairs (Temperature exchange between blue pairs)
7500 step: 2nd dimension among (2↔3), (4↔5), … pairs (Restraint exchange between magenta pairs)
10000 step: 1st dimension among (1↔2), (3↔4), … pairs (Temperature exchange between green pairs)
12500 step: 2nd dimension among (1↔2), (3↔4), … pairs (Restraint exchange between red pairs)
15000 step: 1st dimension among (2↔3), (4↔5), … pairs (Temperature exchange between blue pairs)
:
:
Detailed information about parameter exchange is output in the log file.
If the user uses the following control file (1st and 2nd dimensions were exchanged in the above control file), the exchange pattern is changed:
[REMD]
dimension = 2 # Number of dimension of REMD simulation
exchange_period = 2500 # attempt exchange every 5 ps
type1 = RESTRAINT # restraint function is exchanged
nreplica1 = 6 # number of replicas
rest_function1 = 1 # target restraint function index
type2 = TEMPERATURE # Temperature exchange
nreplica2 = 8 # Number of replicas
parameters2 = 300.00 304.54 309.14 313.80 318.51 323.28 328.10 332.99
2500 step: 2nd dimension among (1↔2), (3↔4), … pairs (Temperature exchange between green pairs)
5000 step: 1st dimension among (2↔3), (4↔5), … pairs (Restraint exchange between magenta pairs)
7500 step: 2nd dimension among (2↔3), (4↔5), … pairs (Temperature exchange between blue pairs)
10000 step: 1st dimension among (1↔2), (3↔4), … pairs (Restraint exchange between red pairs)
12500 step: 2nd dimension among (1↔2), (3↔4), … pairs (Temperature exchange between green pairs)
15000 step: 1st dimension among (2↔3), (4↔5), … pairs (Restraint exchange between magenta pairs)
:
:
