12.3 gREST simulation of trialanine in water
Contents
In this tutorial, we illustrate how to perform and analyze the replica exchange with solute tempering (REST) simulations for alanine tri-peptide. REST was originally developed by Berne et al.[1, 2] to improve the performance of temperature replica exchange MD (T-REMD) simulations. Instead of increase the temperature of the whole system, temperature of the “solute” region is virtually increased. This modification significantly reduces the required number of replicas. In GENESIS, new scheme of REST, referred to as generalized REST (gREST) [3], is implemented. In gREST, the solute region is defined as a part of a molecule and/or a part of the potential energy terms. The rational choice of the solute region in the gREST framework effectively limits the energy space covered in REST and reduces the number of required replica. In the previous REMD tutorial, 20 replicas were needed while in this tutorial only 4 replicas are needed to cover the same temperature range.
When the temperature of the system (= target temperature) and the solute temperature are \(T_0\) and \(T_m\), respectively, the energy in gREST is modified as follows:
\( \displaystyle \beta_0 U_{m} = \beta_0 \left[ \frac{\beta_m}{\beta_0} U_{uu} + \left( \frac{\beta_m}{\beta_0} \right)^{l/n} U_{uv} + U_{vv} \right]\),
where \(\beta_0\) and \(\beta_m\) are the inverse temperatures, \(u\) and \(v\) in the subscript represent the solute and solvent regions, respectively, and \(U_{uv}\) is the interaction between the solute and solvent. For example, the dihedral energy term can be used as the solute. Please note that in the temperature-REMD, each replica has the different temperature of the system, while in gREST all replicas have the same temperature of the system (= \(T_0\)). Only the potential energy (\(U_m\)) is changed by scaling the energy terms in the solute.
Preparation
First, download the tutorial file(tutorial22-12.3). This tutorial consists of five steps: 1) system setup, 2) energy minimization and pre-equilibration, 3) equilibration, 4) production run, and 5) trajectory analysis. Control files for GENESIS are already included in the download file. Since we use the CHARMM force field, we make a symbolic link to the CHARMM toppar directory (see Tutorial 2.2).
# Download the tutorial file $ cd Tutorials $ mv ~/Downloads/tutorial22-12.3.zip ./ $ unzip tutorial22-12.3.zip $ cd tutorial-12.3 $ ls 1_setup/ 2_minimize_pre-equi/ 3_equilibrate/ 4_production/ 5_analysis/ # Make a symbolic link to the CHARMM toppar directory (see Tutorial 2.2) $ ln -s ../../Data/Parameters/toppar_c36_jul21 ./toppar # Setup the directory for binary files of GENESIS 2.0 $ export GENESIS_BIN_DIR=../../../GENESIS_Tutorials-2022/Programs/genesis-2.0/bin/
1. Setup
To setup the system, please follow the steps in the basic tutorial (see Tutorial 3.2). We use the same input PDB and PSF files as in Tutorial 3.2.
# Prepare the input files
$ cd 1_setup
$ ln -s ../../tutorial-3.2/1_setup/3_solvate/wbox.pdb ./
$ ln -s ../../tutorial-3.2/1_setup/3_solvate/wbox.psf ./
2. Minimization and pre-equilibration
Non-physical steric clashes or non-equilibrium geometries in the initial structure must be resolved before the gREST simulation. Here, we do this via five steps: 1) minimization (step2.1), 2) equilibration in NVT ensemble (step2.2), 3) relaxation of the simulation box in NPT ensemble with positional restraints (step2.3), 4) equilibration in NPT ensemble without positional restraints, and 5) equilibration in NPT ensemble with 3.5 fs time step and r-RESPA (step2.5). In the 2_minimize_pre-equil directory, all control files “step2.*.inp” are given. For further details of the control file and choice of parameters, please refer to basic tutorials section (see Tutorial 3.2) and Tutorial 10.1 for simulations with large time step.
Let’s change directory.
#Change directory to 2_minimize_pre-equi $ cd ../2_minimize_pre-equi $ ls step2.1.inp step2.2.inp step2.3.inp step2.4.inp step2.5.inp
2.1. Minimization
The first step of the simulation is to run energy minimization of the system, in order to remove atomic clashes in the initial structure. The control file “step2.1.inp” is as follows:
[INPUT] topfile = ../toppar/top_all36_prot.rtf # topology file parfile = ../toppar/par_all36m_prot.prm # parameter file strfile = ../toppar/toppar_water_ions.str # stream file psffile = ../1_setup/wbox.psf # protein structure file pdbfile = ../1_setup/wbox.pdb # PDB file [OUTPUT] dcdfile = step2.1.dcd # DCD trajectory file rstfile = step2.1.rst # restart file [ENERGY] forcefield = CHARMM # [CHARMM] electrostatic = PME # [PME] switchdist = 10.0 # switch distance cutoffdist = 12.0 # cutoff distance pairlistdist = 13.5 # pair-list distance vdw_force_switch = YES # force switch option for van der Waals contact_check = YES # check atomic clash [MINIMIZE] method = SD # [SD] nsteps = 2000 # number of minimization steps eneout_period = 50 # energy output period crdout_period = 50 # coordinates output period rstout_period = 2000 # restart output period [BOUNDARY] type = PBC # [PBC] box_size_x = 50.2 # box size (x) in [PBC] box_size_y = 50.2 # box size (y) in [PBC] box_size_z = 50.2 # box size (z) in [PBC] [SELECTION] group1 = sid:PROA and heavy # restraint group 1 [RESTRAINTS] nfunctions = 1 # number of functions function1 = POSI # restraint function type direction1 = ALL # direction [ALL,X,Y,Z] constant1 = 1.0 # force constant select_index1 = 1 # restrained groups
To execute the calculation, use the following command:
# Run energy minimization $ export OMP_NUM_THREADS=5 $ mpirun -np 8 $GENESIS_BIN_DIR/spdyn step2.1.inp > step2.1.out
After the calculation, check the trajectory by using VMD. We can see that the atoms are slightly moved, but the atomic clashes are actually removed. In this step, we used 8 MPI processors and 5 OpenMP threads, namely, 40 CPU cores.
2.2. Equilibration of the system at 300 K
The second step of the simulation is to heat up the system, with restraint on the peptide heavy atoms, to 300 K. The heating is performed via annealing process wherein the temperature is increased by 3 K every 500 steps. Total simulation is 100 ps. Note that in this step the integrator is Velocity Verlet (VVER) and Bussi thermostat. Please view the control input file (step2.2.inp). The following is the important options in the control file.
[DYNAMICS] integrator = VVER # [LEAP,VVER] nsteps = 50000 # number of MD steps timestep = 0.002 # timestep (ps) eneout_period = 500 # energy output period crdout_period = 500 # coordinates output period rstout_period = 50000 # restart output period annealing = YES # simulated annealing anneal_period = 500 # annealing period dtemperature = 3 # temperature change at annealing (K) iseed = 31415 # random number seed [CONSTRAINTS] rigid_bond = YES # constraints all bonds involving hydrogen [ENSEMBLE] ensemble = NVT # [NVE,NVT,NPT,NPAT,NPgT] tpcontrol = BUSSI # [NO,BERENDSEN,BUSSI,NHC] temperature = 0.1 # initial and target temperature (K)
To execute the simulation, we use similar commands as previous step.
# Run heating step $ export OMP_NUM_THREADS=5 $ mpirun -np 8 $GENESIS_BIN_DIR/spdyn step2.2.inp > step2.2.out
2.3. Relaxation of the simulation box
After the system reached the desired temperature 300.00 K, we relax the simulation box in NPT ensemble at 300 K and 1 atm for 50 ps. The following is the important options in the control file (step2.3.inp).
[DYNAMICS] integrator = VVER # [LEAP,VVER] nsteps = 25000 # number of MD steps timestep = 0.002 # timestep (ps) eneout_period = 500 # energy output period crdout_period = 500 # coordinates output period rstout_period = 25000 # restart output period [ENSEMBLE] ensemble = NPT # [NVE,NVT,NPT,NPAT,NPgT] tpcontrol = BUSSI # [NO,BERENDSEN,BUSSI,NHC] temperature = 300 # initial and target temperature (K) pressure = 1.0 # target pressure (atm)
To execute the simulation, we use the following command.
# Run relaxation of the system with restraints $ export OMP_NUM_THREADS=5 $ mpirun -np 8 $GENESIS_BIN_DIR/spdyn step2.3.inp > step2.3.out
2.4. Equilibration with no positional restraints
We equilibrate all atom positions including the peptide atoms without positional restraints (removes [RESTRAINTS]section). Here we turn on the functions of hydrogen mass repartitioning (HMR) and group temperature/pressure (group T/P) to use the 3.5 fs time step in the next equilibration step. Please note that this option is only available in GENESIS v2.0 and later. The following is the important options in the control file (step2.4.inp).
[DYNAMICS] integrator = VVER # [LEAP,VVER,VRES] nsteps = 25000 # number of MD steps (50 ps) timestep = 0.002 # timestep (2 fs) eneout_period = 500 # energy output period crdout_period = 500 # coordinates output period rstout_period = 25000 # restart output period hydrogen_mr = yes # Turn on HMR hmr_ratio = 3.0 hmr_ratio_xh1 = 2.0 hmr_target = solute # Apply HMR only to solute [ENSEMBLE] ensemble = NPT # [NVE,NVT,NPT,NPAT,NPgT] tpcontrol = BUSSI # [NO,BERENDSEN,BUSSI,LANGEVIN] temperature = 300 # initial and target temperature (K) pressure = 1.0 # target pressure (atm) group_tp = YES # usage of group tempeature and pressure
To run this step, we use the following command.
# Run equilbration without restraints
$ export OMP_NUM_THREADS=5
$ mpirun -np 8 $GENESIS_BIN_DIR/spdyn step2.4.inp > step2.4.out
2.5. Equilibration with 3.5 fs time step
Finally we run an additional 105-ps pre-equilibration simulation in NPT ensemble. In this step, we equilibrate the system with the multiple time step integrator, r-RESPA (VRES, timestep of 3.5 fs). The integration with the longer time step requires HMR and group T/P options. The following is the important options in the control file (step2.5.inp).
[DYNAMICS] integrator = VRES # [LEAP,VVER,VRES] nsteps = 30000 # number of MD steps (105 ps) timestep = 0.0035 # timestep (3.5 fs) eneout_period = 300 # energy output period (1.05 ps) crdout_period = 300 # coordinates output period (1.05 ps) rstout_period = 30000 # restart output period nbupdate_period = 6 # nonbond update period elec_long_period = 2 # period of reciprocal space calculation thermostat_period = 6 # period of thermostat update barostat_period = 6 # period of barostat update hydrogen_mr = yes # Turn on HMR hmr_ratio = 3.0 hmr_ratio_xh1 = 2.0 hmr_target = solute # Apply HMR only to solute [ENSEMBLE] ensemble = NPT # [NVE,NVT,NPT,NPAT,NPgT] tpcontrol = BUSSI # [NO,BERENDSEN,BUSSI,LANGEVIN] temperature = 300 # initial and target temperature (K) pressure = 1.0 # target pressure (atm) group_tp = YES # usage of group tempeature and pressure
To run this step, we use the following command.
# Run equilbration without restraints
$ export OMP_NUM_THREADS=5
$ mpirun -np 8 $GENESIS_BIN_DIR/spdyn step2.5.inp > step2.5.out
3. Equilibration for gREST
In this step, we equilibrate the system for each replica without replica exchange. Let’s move to the directory for equilibration of gREST.
#Change directory to 3_equilibrate $ cd ../3_equilibrate $ ls step3.inp
The control file of gREST is very similar to that of T-REMD. The only difference is that gREST needs select_indexN and param_typeN to specify the “solute” region. The rest of system is regarded as “solvent”. In this example, we define the the LJ and dihedral angle of potential energy terms in Ala3 molecule as the solute region (param_type1 = D L in [REMD] section and group1 = ai:1-42 in [SELECTION] section). In default, all of potential energy terms are treated as the solute (param_type1 = ALL in [REMD] section). If you want to treat other parts of potential energy terms as the solute, param_typeN parameter may be modified (e.g. param_type1 = C CM to specify only charge and CMAP terms as solute.).
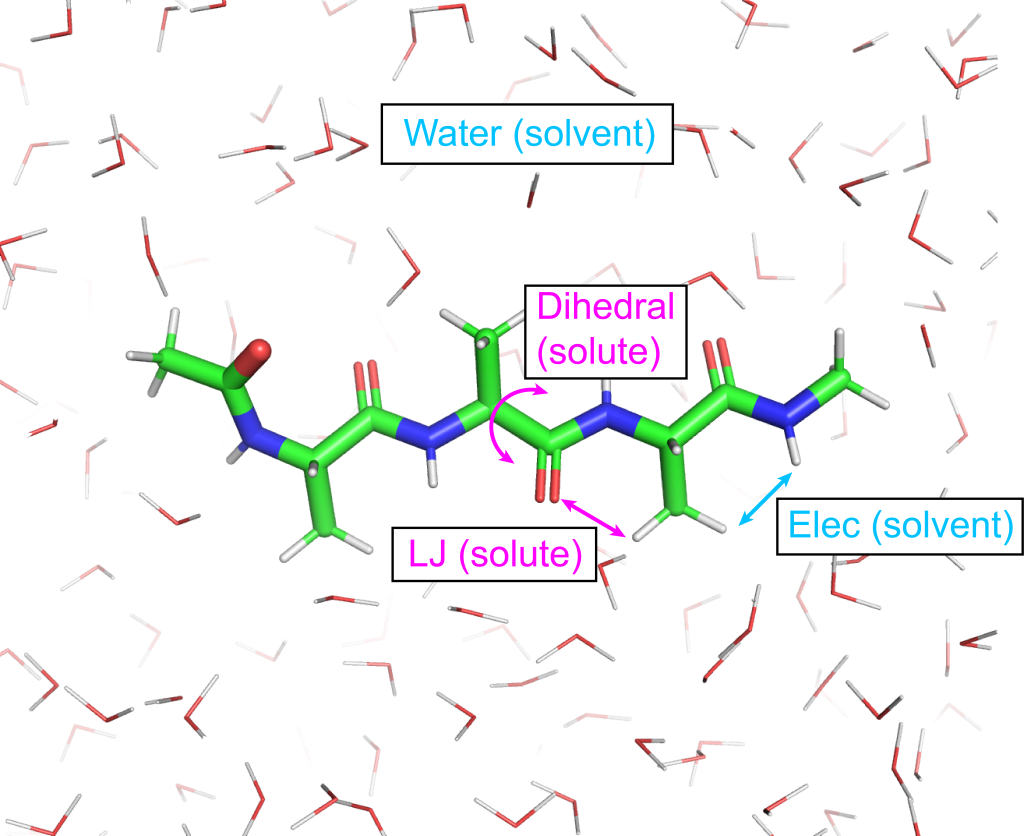
We here use four replicas with the solute temperature range of 300.00 K – 351.26 K (parameters1 = 300.00 318.12 337.14 351.26 in [REMD] section) as a example. If you apply gREST to your system, you must determine the number and the temperatures of replicas. To find the number of replicas and temperatures, please refer to the appendix ((see Automatic Parameter Tuning for REMD, REUS, REST)). Each replica must be equilibrated at the selected temperature just like conventional MD simulations. Note that in this step as well as the next step (production run) we use NVT ensemble. The r-RESPA integrator is combined with HMR and group T/P in order to use 3.5 fs time step. The following is the important options in the control file (step3.inp).
[REMD] dimension = 1 # number of parameter types exchange_period = 0 # NO exchange for equilibration iseed = 3141592 type1 = REST # Replica Exchange with Solute Tempering nreplica1 = 4 # number of replicas parameters1 = 300.00 318.12 337.14 351.26 # list of solute temperatures select_index1 = 1 # solute region selection param_type1 = D L # function types # valid keywords are: # ALL (default), BOND(B), ANGLE(A), # DIHEDRAL(D), IMPROPER(I), CMAP(CM), # CHARGE(C), LJ(L)... # See manual for further details. [SELECTION] group1 = ai:1-42 [DYNAMICS] integrator = VRES # [LEAP,VVER,VRES] nsteps = 30000 # number of MD steps (105ps) timestep = 0.0035 # timestep (3.5fs) eneout_period = 300 # energy output period (1.05ps) crdout_period = 300 # coordinates output period (1.05ps) rstout_period = 3000 # restart output period nbupdate_period = 6 # nonbond update period elec_long_period = 2 # period of reciprocal space calculation thermostat_period = 6 # period of thermostat update barostat_period = 6 # period of barostat update hydrogen_mr = yes # Turn on HMR hmr_ratio = 3.0 hmr_ratio_xh1 = 2.0 hmr_target = solute # Apply HMR only to solute [ENSEMBLE] ensemble = NVT # [NVE,NVT,NPT] tpcontrol = BUSSI # thermostat and barostat temperature = 300.0 # initial temperature (K) pressure = 1.0 # target pressure (atm) group_tp = YES
The following command performs a 105 ps NVT gREST simulations. Here we use 160 (= 32 MPI x 5 OpenMP) CPU cores for the simulations.
# Run gREST equilibiration step $ export OMP_NUM_THREADS=5 $ mpirun -np 32 $GENESIS_BIN_DIR/spdyn step3.inp > step3.out
4. Production
Since we have now completed all preparation steps, now we can start running the production simulation. Let’s move to the directory for production of gREST.
#Change directory $ cd ../4_production $ ls step4.inp
We run a short simulation for 10.5 ns for the purpose of tutorial, however longer simulation might be needed for conversion. The following control file is used to run the simulation in NVT ensemble. One needs to modify the control file for the equilibration such that the exchange_period has a non-zero value (e.g. 3000 steps (10.5 ps)) and [INPUT] and [OUTPUT] files are properly refered. In order to analyze the free energy, you need to select the analysis_grest=YES in [REMD]. The option enables calculation of energies with different solute temperatures. The energies with different solute temperatures of each replica are written in step4_rep{}.ene. The important part of the control file (step4.inp) is as follows:
[REMD] dimension = 1 # number of parameter types exchange_period = 3000 # exchange per 3000*3.5fs = 10.5 ps type1 = REST # Replica Exchange with Solute Tempering nreplica1 = 4 # number of replicas parameters1 = 300.00 318.12 337.14 351.26 # list of solute temperatures select_index1 = 1 # solute region selection param_type1 = D L # function types # valid keywords are: # ALL (default), BOND, ANGLE, # DIHEDRAL, IMPROPER, CMAP, # CHARGE LJ... # See manual for further details. analysis_grest = YES [SELECTION] group1 = ai:1-42 [DYNAMICS] integrator = VRES # [LEAP,VVER,VRES] nsteps = 3000000 # number of MD steps (10.5 ns) timestep = 0.0035 # timestep (3.5 fs) eneout_period = 300 # energy output period (1.05 ps) crdout_period = 300 # coordinates output period (1.05 ps) rstout_period = 30000 # restart output period (105 ps) nbupdate_period = 6 # nonbond update period elec_long_period = 2 # period of reciprocal space calculation thermostat_period = 6 # period of thermostat update barostat_period = 6 # period of barostat update hydrogen_mr = yes # Turn on HMR hmr_ratio = 3.0 hmr_ratio_xh1 = 2.0 hmr_target = solute # Apply HMR only to solute [ENSEMBLE] ensemble = NVT # [NVE,NVT,NPT] tpcontrol = BUSSI # thermostat and barostat temperature = 300.00 # K group_tp = YES # usage of group tempeature and pressure
To run gREST production run, we use the following commands.
# Run gREST production step $ export OMP_NUM_THREADS=5 $ mpirun -np 32 $GENESIS_BIN_DIR/spdyn step4.inp > step4.out
5. Analysis
In this tutorial, we mainly focus on calculating PMF of the end to end distance distribution at 300 K. In which, we use all temperatures trajectory upon applying the Multistate Bennett Acceptance Ratio (MBAR) re-weighting method. For information on MBAR method, please check [4]. However, before calculating PMF, we first check the simulation by calculating acceptance ratio, replica random walk and temperature potential energy distribution.
In gREST control file, we setup the exchange_period=3000 which means replica exchange is attempt every 10.5 ps. In the log output of the gREST simulation, we can see the information about replica-exchange attempts at every exchange_period steps.
REMD> Step: 2982000 Dimension: 1 ExchangePattern: 1 Replica ExchangeTrial AcceptanceRatio Before After 1 4 > 3 A 400 / 497 351.260 337.140 2 2 > 1 A 394 / 497 318.120 300.000 3 1 > 2 A 394 / 497 300.000 318.120 4 3 > 4 A 400 / 497 337.140 351.260 Parameter : 337.140 300.000 318.120 351.260 RepIDtoParmID: 3 1 2 4 ParmIDtoRepID: 2 3 1 4
In this log file, we should pay attention to the AcceptanceRatio values. If those values are much lower than target, you should review the parameters of your simulation, such as modifying the temperature range. In this table, 'A' and 'R' mean that the exchange at this step is accepted or rejected, respectively. The last two columns show replica temperatures before and after the exchange trials, respectively.
Lines in red summarize the locations and parameters after replica exchanges. The Parameter line gives the temperature of each replica in gREST simulation. The RepIDtoParmID line stands for the permutation function that converts Replica ID to Parameter ID. For example, in the 1st column, 3 is written, which means that the temperature of Replica 1 is set to 337.140 K. The ParmIDtoRepID line also represents the permutation function that converts Parameter ID to Replica ID. For example, in the 3th column, 1 is written, which means that Parameter 3 (corresponding to the replica temperature, 337.140 K) is located in Replica 1.
Now, please change the directory to analysis and proceed with the following steps:
# change directory
$ cd ../5_analysis
$ ls
1_calc_ratio 2_plot_index 3_sort 4_clac_dist 5_mbar 6_pmf
5.1. Calculate the acceptance ratio of each replica
Acceptance ratio of replica exchange is one of the important factors that determine the efficiency of gREST simulations. The acceptance ratio is displayed in a standard log output “step4.out“, and we examine the data from the last step. Here, we show an example how to examine the data. Note that the acceptance ratio of replica “A” to “B” is identical to “B” to “A”, and thus we calculate only “A” to “B”. For this calculation, you can use the script “calc_ratio.sh“.
# change directory
$ cd 1_calc_ratio
$ ls
calc_ratio.sh
# make the file executable and use it
$ chmod u+x calc_ratio.sh
$ ./calc_ratio.sh
1 > 2 0.826
2 > 3 0.804
3 > 4 0.894
The file “calc_ratio.sh” contains the following commands:
# get acceptance ratios between adjacent parameter IDs
$ grep " 1 > 2" ../../4_production/step4.out | tail -1 > acceptance_ratio.dat
$ grep " 2 > 3" ../../4_production/step4.out | tail -1 >> acceptance_ratio.dat
$ grep " 3 > 4" ../../4_production/step4.out | tail -1 >> acceptance_ratio.dat
# calculate the ratios
$ awk '{print $2,$3,$4,$6/$8}' acceptance_ratio.dat
Note that the average acceptance ration in this case is quite high, representing that a much larger temperature range can be achieved with 4 replica. For consistency and comparison to REMD tutorial, the temperature range was kept the same.
5.2. Plot time courses of replica indices and temperatures
To examine the random walks of each replica in temperature space, we analyze time course of the replica indices. We need to plot the values of the “ParmIDtoRepID” lines from step4.out for a chosen starting replica temperature, for example 300 K (first column), versus time. Using following commands, we can get replica IDs in each snapshot.
# change directory
$ cd ../2_plot_index
$ ls
plot_index.sh plot_temperature.sh
# make the file executable and use it
$ chmod u+x plot_index.sh
$ ./plot_index.sh
The file “plot_index.sh” contains the following commands:
# get replica IDs in each snapshot
$ grep "ParmIDtoRepID:" ../../4_production/step4.out | sed 's/ParmIDtoRepID:/ /' > param.dat
Using following gnuplot commands, we can plot the replica IDs in each snapshot.
# make input file for gnuplot
cat << EOF > tmp.plt
set terminal png
set yrange [0:5]
set mxtics
set mytics
set xlabel "Time (ns)"
set ylabel "Replica ID"
# plot replica IDs which visited 300.00 K
set output "output_index1.png"
plot "param.dat" using (\$0*10.5/1000):1 with points pt 5 ps 0.5 lt 1 title "300.00 K"
# plot replica IDs which visited 318.12 K
set output "output_index2.png"
plot "param.dat" using (\$0*10.5/1000):2 with points pt 5 ps 0.5 lt 1 title "318.12 K"
# plot replica IDs which visited 337.14 K
set output "output_index3.png"
plot "param.dat" using (\$0*10.5/1000):3 with points pt 5 ps 0.5 lt 1 title "337.14 K"
# plot replica IDs which visited 351.26 K
set output "output_index4.png"
plot "param.dat" using (\$0*10.5/1000):4 with points pt 5 ps 0.5 lt 1 title "351.26 K"
EOF
# execute gnuplot
gnuplot ./tmp.plt
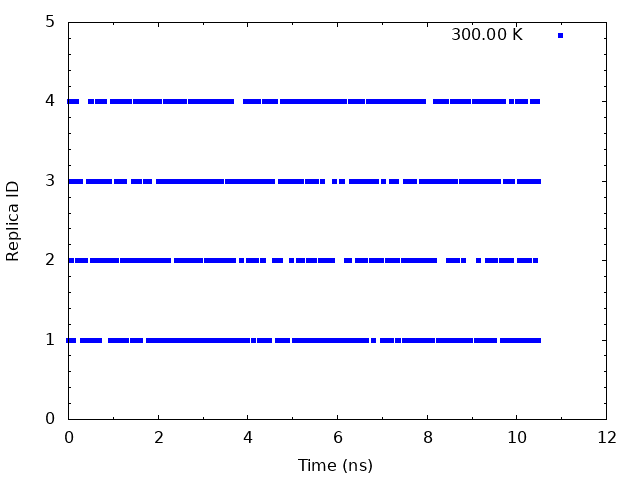
This graph indicate that the temperatures (300 K) visit randomly each replica, and thus random walks in the temperature spaces are successfully realized.
We also plot time courses of temperatures in one replica. We need to plot one column in the “Parameter :” lines in step4.out versus time. Using following commands. we can get replica temperatures in each snapshot.
# make the file executable and use it
$ chmod u+x plot_temperature.sh
$ ./plot_temperature.sh
The file “plot_temperature.sh” contains the following commands:
# get replica temperatures in each snapshot
$ grep "Parameter :" ../../4_production/step4.out | sed 's/Parameter :/ /' > temperature.dat
As previous step, we can use gunplot script to plot the parameter IDs (= replica temperatures) in each snapshot.
# make input file for gnuplot cat << EOF > tmp.plt set term png set mxtics set mytics set xlabel "Time (ps)" set ylabel "Temperature (K)" set output "output_temperature_rep1.png" plot "temperature.dat" using (\$0*10.5/1000):1 with lines lc rgb "blue" title "repID=1 " set output "output_temperature_rep2.png" plot "temperature.dat" using (\$0*10.5/1000):2 with lines lc rgb "blue" title "repID=2 " set output "output_temperature_rep3.png" plot "temperature.dat" using (\$0*10.5/1000):3 with lines lc rgb "blue" title "repID=3 " set output "output_temperature_rep4.png" plot "temperature.dat" using (\$0*10.5/1000):4 with lines lc rgb "blue" title "repID=4 " EOF # execute gnuplot gnuplot tmp.plt

The temperatures of each replica during the simulation are distributed in all temperatures assigned. It means that correct annealing of the system is realized.
5.3. Sort coordinates in DCD trajectory files by parameters
The temperature in output DCD files of gREST simulation have all range of temperatures, due to the exchange. Therefore, to analyze the simulation further, we first need to sort the frames in the trajectory based on their temperature. To do that, we use GENESIS analysis tool (remd_convert). Sorting is done based on the information written in remfiles generated from the gREST simulation. Concomitantly, we also sort log and energy files for each replica based on temperature parameters. The sorted ene files will be used in the next step in MBAR analysis.
# change directory
$ cd ../3_sort
$ ls
remd_convert.inp
# Sort frames by parameters
$ $GENESIS_BIN_DIR/remd_convert remd_convert.inp | tee remd_convert.out
This example sorts the trajectory of the solute temperature of 300.00 K (parameterID = 1) (convert_type = PARAMETER, convert_ids = 1 in [OPTION] section). The system is aligned to the backbone of central alanine during the sorting (fitting_atom = 2 in [FITTING] section, group2 = resno:2 and (an:N or an:CA or an:C or an:O in [SELECTION] section), and the water molecules are removed from the sorted trajectory (trjout_atom = 1 in [OPTION] section, group1 = ai:1-42 in [SELECTION] section). The control file “remd_convert.inp” is as follows:
[INPUT] psffile = ../../1_setup/wbox.psf reffile = ../../1_setup/wbox.pdb dcdfile = ../../4_production/step4_rep{}.dcd # see remd_conv.sh remfile = ../../4_production/step4_rep{}.rem # see remd_conv.sh enefile = ../../4_production/step4_rep{}.ene # enefile of gREST [OUTPUT] pdbfile = param.pdb # pdbfile trjfile = param{}.dcd # sorted dcdfile enefile = param{}.ene # sorted enefile [SELECTION] group1 = ai:1-42 # only tri-alanine group2 = resno:2 and (an:N or an:CA or an:C or an:O) # backbone of 2nd ALA [FITTING] fitting_method = TR+ROT # center-of-mass translation + rotation fitting fitting_atom = 2 # fitting to backbone of central alanine mass_weight = YES # mass-weighted fitting [OPTION] check_only = NO convert_type = PARAMETER convert_ids = # only lowest T replicas num_replicas = 4 nsteps = 3000000 exchange_period = 3000 crdout_period = 300 # trjectory output frequency in REMD eneout_period = 300 # energy output frequency in REMD trjout_format = DCD trjout_type = COOR+BOX trjout_atom = 1 # output only tri-alanine moiety pbc_correct = NO # nothing will happen when water mols excluded
Now we have sorted temperature log, ene and DCD file which will be used in the following analysis steps.
One can check the sorted trajectory by following commands:
# Check the sorted dcd file for ParameterID 1 (300.00 K)
$ vmd ./param.pdb ./param1.dcd
“param*.ene” files contain the energies at different solute temperatures at the snapshots of each replica. For example, param1.ene is shown below. The second column represents the potential energy at 300.00 K. The third, fourth, and fifth columns represent the potential energies, which are estimated at 318.12 K, 337.14 K, and 351.26 K, respectively, using the trajectory of 300.00 K.
300 -38420.8021 -38420.7701 -38420.6700 -38420.5615 600 -38437.8129 -38437.7333 -38437.5876 -38437.4485 900 -38454.1105 -38454.6977 -38455.1828 -38455.4708 1200 -38523.4070 -38523.3613 -38523.2382 -38523.1079 1500 -38452.2631 -38452.5241 -38452.6885 -38452.7519 1800 -38472.7160 -38472.7599 -38472.7295 -38472.6667
5.4. Calculating end to end distance
In order to calculate the potential of the mean force (PMF) of the end-to-end distance distribution, in the current subsection we calculate the distance between the two terminal alanine (OY_HNT).
# change directory
$ cd ../4_calc_distance
$ ls
calc_dist.sh
# make the file executable and execute the script
$ chmod u+x calc_dist.sh
$ ./calc_dist.sh
In the script “calc_dist.sh”, we use GENESIS analysis tool trj_analysis as follow:
for i in 1 2 3 4; do # Make input files for each Parameter ID. cat << EOF > inp${i} [INPUT] reffile = ../3_sort/param.pdb [OUTPUT] disfile = param${i}.dis [TRAJECTORY] trjfile1 = ../3_sort/param${i}.dcd md_step1 = 3000000 # number of MD steps mdout_period1 = 300 # MD output period ana_period1 = 300 # analysis period repeat1 = 1 trj_format = DCD # (PDB/DCD) trj_type = COOR+BOX # (COOR/COOR+BOX) trj_natom = 0 # (0:uses reference PDB atom count) [OPTION] check_only = NO # (YES/NO) distance1 = PROA:1:ALA:OY PROA:3:ALA:HNT EOF # Execute trj_analysis trj_analysis ./inp${i} done
5.5. MBAR analysis
In order to use conformers from temperatures higher than the target temperature (300K), we use the MBAR method. MBAR reweights each snapshot of each replica into the target temperature and provide the unbiased weight for each snapshot:
\( \displaystyle W_{jn} = \frac{1}{c} \frac{\exp [-\beta_0 U_0(x_{jn})]}{\sum_k N_k \exp [\beta_0 (f_k – U_k(x_{jn}))]},\)
where \(x_{jn}\) is a configuration of snapshot \(n\) at replica \(j\), \(f_k\) is the free energy of replica \(k\), and \(c\) is a normalization constant. \(U_0\) is the potential energy function at \(\beta_0\). \(U_k(x_{jn})\) corresponds to energies in param*.ene files. Please note that the values of the solute temperatures are not required for reweighting because their information is already included in \(U_k(x_{jn})\).
We apply GENESIS mbar_analysis tool where we use our sorted energy files as cv.
# change directory
$ cd ../5_mbar
$ ls
mbar_analysis.inp
# We run the analysis using the following command.
$ $GENESIS_BIN_DIR/mbar_analysis mbar_analysis.inp | tee mbar_analysis.log
The control file ” mbar_analysis.inp” is as follows:
[INPUT] cvfile = ../3_sort/param{}.ene # input cv file [OUTPUT] fenefile = fene.dat weightfile = weight{}.dat [MBAR] nreplica = 4 input_type = REST dimension = 1 nblocks = 1 temperature = 300.00 target_temperature = 300.00
input_type is set to REST to reweight gREST trajectories. temperature and target_temperature are set to 300.00. As explained above, the information about solute temperatures are not needed for reweighting. mbar_analysis produces “fene.dat” file containing the evaluated relative free energies and 4 “weight*.dat” files containing the weights of each snapshot for each replica, which are reweighted to 300.00 K. For example, weight1.dat is shown below. The first line corresponds to the weight of the first snapshot in param1.dcd. Each weight represents the probability of each snapshot at 300.00 K.
$ head weight1.dat 300 1.836119160862883E-005 600 1.962571423551713E-005 900 4.720421550879687E-006 1200 1.883861114197157E-005 1500 1.100339116123613E-005 1800 1.636948804025372E-005 2100 1.812549281313518E-005 2400 2.113452205982935E-005 2700 8.409791663698055E-006 3000 1.536017798686323E-005
5.6. Calculating PMF of distance distribution
The final step of this tutorial is to use the calculated distances in 5.4. and weight files from MBAR analysis (5.5.) to calculate the PMF of the end-to-end distance distribution in Ala3. We use another tool in GENESIS (pmf_analysis) as follow:
# change directory
$ cd ../6_pmf
$ ls
pmf_analysis.inp plot_pmf.sh
# We run the analysis using the following command:
$ $GENESIS_BIN_DIR/pmf_analysis pmf_analysis.inp | tee pmf_analysis.log
The control file ” pmf_analysis.inp” is as follows:
[INPUT] cvfile = ../4_calc_dist/param{}.dis weightfile = ../5_mbar/weight{}.dat [OUTPUT] pmffile = dist.pmf [OPTION] nreplica = 4 # number of replicas dimension = 1 # dimension of cv space temperature = 300.0 grids1 = 0 15 101 # (min max num_of_bins) band_width1 = 0.1 is_periodic1 = NO # periodicity of cv1
We plot the PMF using the provided script, “plot_pmf.sh”.
# make the file executable and plot PMF
$ chmod u+x plot_pmf.sh
$ ./plot_pmf.sh
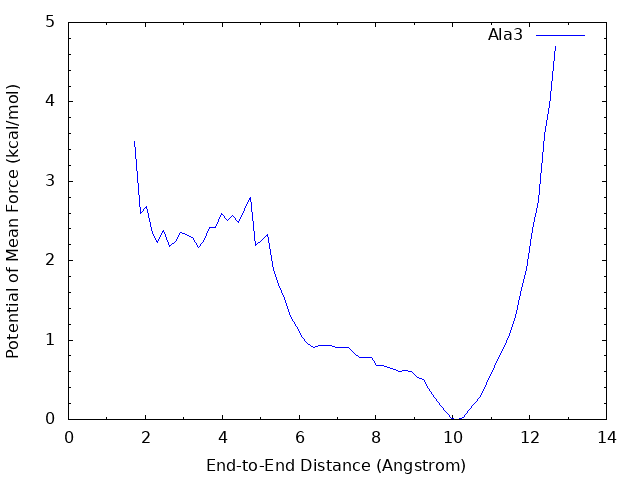
We can see that there is the global energy minimum around r = 10 Å and a local energy minimum around r = 2.8 Å. The latter corresponds to the α-helix conformation, where the hydrogen bond between OY and HNT is formed. These results suggest that in water the alanine tripeptide tends to form an extended conformation rather than α-helix.
References
- P. Liu, B. Kim, R. A. Friesner, B. J. Berne, Replica exchange with solute tempering: A method for sampling biological systems in explicit water. Proc. Natl. Acad. Sci. U.S.A.102, 13749–13754 (2005).
- L. Wang, R. A. Friesner, B. J. Berne, Replica exchange with solute scaling: A more 683efficient version of replica exchange with solute tempering (REST2). J. Phys. Chem. B 115, 9431–9438 (2011).
- M. Kamiya, Y. Sugita, Flexible selection of the solute region in replica exchange with solute tempering: Application to protein-folding simulations. J. Chem. Phys. 149, 072304 (2018)
- M. Shirts et al., J. Chem. Phys., 129, 124105-124114 (2008).
Written Mar 1, 2018 by Motoshi Kamiya@RIKEN Computational biophysics research team
Updated Feb 25, 2019 by Yasuhiro Matsunaga@RIKEN R-CCS
Updated Aug 31, 2019 by Suyong Re@RIKEN BDR
Updated Jul 2, 2020 by Chigusa Kobayashi@RIKEN R-CCS
Updated Jul 1, 2022 by Hisham Dokainish@RIKEN CPR
Updated Sep 14, 2022 by Hiraku Oshima@RIKEN BDR
