sasa_analysis
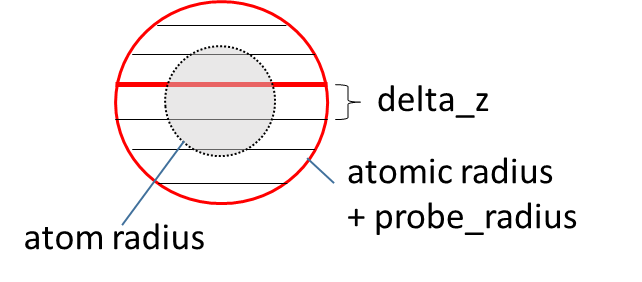
sasa_analysis calculates solvent accessible surface area (SASA) of the solute. Atomic SASA is calculated for each selected atom in the solute molecule, and the summation of those atomic SASA is output as total SASA. In this program, each selected atom is approximated by the set of the circles, with certain spacing delta_z. The solvent probe radius is determined by probe_radius. The radius of selected atoms is determined by additional input file radi_file, in which atom types and their radius are listed. The cell size (box_size_x,y,z/num_cells_x,y,z) should be larger than (max atom radius + probe_radius) x 2. According to this rule, cell size usually becomes larger than 8.0Å. output_style = history outputs only the time history of the total SASA, output_style = atomic outputs the time-averaged atomic SASA over the analysis frames, and output = atomic+history outputs the time history of both total and atomic SASA.
(EX1) SASA calculation for a BPTI
This input calculates the SASA of a protein BPTI (group1) in water box with PBC.
[INPUT] psffile = ionize.psf reffile = ionize.pdb pdbfile = ionize.pdb [OUTPUT] txtfile = BPTI.out [TRAJECTORY] trjfile1 = run.dcd md_step1 = 500000 mdout_period1 = 5000 ana_period1 = 5000 repeat1 = 1 trj_format = DCD trj_type = COOR+BOX trj_natom = 0 [BOUNDARY] type = PBC box_size_x = 68.25815 box_size_y = 80.24045 box_size_z = 66.58892 domain_x = 2 domain_y = 2 domain_z = 2 num_cells_x = 8 num_cells_y = 10 num_cells_z = 8 [ENSEMBLE] ensemble = NPT [SELECTION] group1 = ai:1-892 [SPANA_OPTION] buffer = 8 wrap = yes box_size = TRAJECTORY [SASA_OPTION] solute = 1 radi_file = radi_list probe_radius = 1.4 delta_z = 0.2 output_style = atomic+history recenter = 1
(EX2) Total SASA calculation for multiple villins under the crowding condition
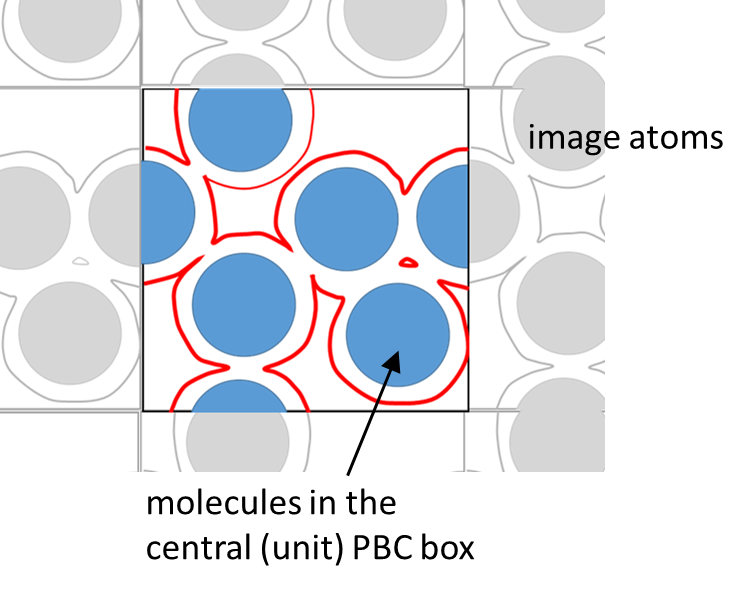
This example calculates the total SASA for multiple villins under the crowding condition. When boundary type is PBC, sasa_analysis takes account the image atoms around the unit box. However, the atomic SASA (and their summation) only for those atoms in the unit PBC box is output.
[INPUT] psffile = villin_crowding.psf reffile = villin_crowding.pdb pdbfile = villin_crowding.pdb [OUTPUT] txtfile = villin.out [TRAJECTORY] trjfile1 = prod.dcd md_step1 = 500000 mdout_period1 = 5000 ana_period1 = 5000 repeat1 = 1 trj_format = DCD trj_type = COOR+BOX [BOUNDARY] type = PBC # [PBC] box_size_x = 142 box_size_y = 142 box_size_z = 142 domain_x = 2 domain_y = 2 domain_z = 2 num_cells_x = 14 num_cells_y = 14 num_cells_z = 14 [ENSEMBLE] ensemble = NVT [SELECTION] group1 = ai:1-13112 [SPANA_OPTION] buffer = 10 wrap = yes box_size = TRAJECTORY [SASA_OPTION] solute = 1 radi_file = radi_list probe_radius = 1.4 delta_z = 0.2 output_style = atomic+history
sasa_analysis can consider or ignore the influence of surrounding molecules
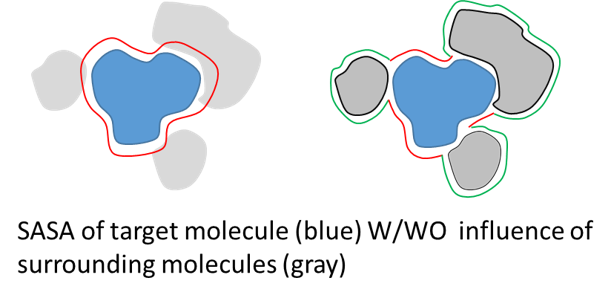 sasa_analysis calculates total and atomic sasa for all groups specified in [SELECTION] section, but outputs only those value for a group specified in option solute in [SASA_OPTION] section. Using the same system as EX2, if you want to calculate SASA for one of the villins, you should select the target molecule as follows. In this case influence of the surrounding molecules are not considered.
sasa_analysis calculates total and atomic sasa for all groups specified in [SELECTION] section, but outputs only those value for a group specified in option solute in [SASA_OPTION] section. Using the same system as EX2, if you want to calculate SASA for one of the villins, you should select the target molecule as follows. In this case influence of the surrounding molecules are not considered.
[SELECTION] group1 = ai:1-596 [SASA_OPTION] solute = 1
If you specify the groups like this, SASA for group1 (a target villin) is calculated taking account the presence of other villins.
[SELECTION] group1 = ai:1-596 group2 = ai:597-13112 [SASA_OPTION] solute = 1