hb_analysis
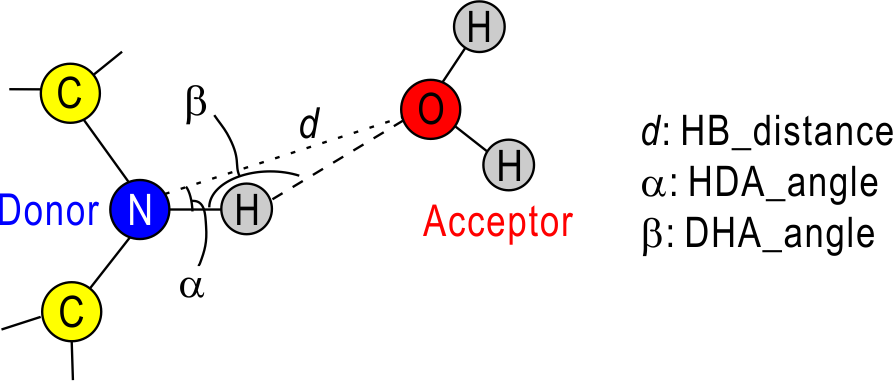
This tool is used to examine H-bonds formed in the selected two atom groups. For the detection of H-bonds, we need the three geometrical parameters: hb_distance (d), HDA_angle (α), and DHA_angle (β). In the default, d = 3.4 Å, α = 30.0 deg. and β = 120.0 deg. If you use your own definition, you can change these values in [OPTION] section. In the current version, H-bonds are examined only for nitrogen or oxygen atoms.
This tool has two calculation options: CountSnap and CountAtom. In this sample page, we illustrate samples for these two options.
case 1: Count the number of H-bonds formed in every snapshot
When output_type = Count_snap, the number of H-bonds formed between ‘analysis_atom‘ and ‘target_atom‘ groups is output per snapshot used for the analysis. When boundary_type = PBC, the examination of H-bonds is done taking account of PBC.
[INPUT]
psffile = BPTI_ionize.psf
reffile = BPTI_ionize.pdb
[OUTPUT]
txtfile = BPTI_internal_water_hbonds.txt
hb_listfile = BPTI_internal_water_hbonds.list
[TRAJECTORY]
trjfile1 = run.dcd
md_step1 = 10
mdout_period1 = 1
ana_period1 = 1
trj_format = DCD
trj_type = COOR+BOX
[SELECTION]
group1 = sid:BPTI & resno:1-58
group2 = resname:TIP3 & resno:2065
[OPTION]
check_only = NO
output_type = Count_Snap # number of H-bonds are output per snapshot
analysis_atom = 1
target_atom = 2
boundary_type = PBC
hb_distance = 3.4
dha_angle = 120.0
hda_angle = 30.0
When ‘hb_listfile‘ name is specified in [OUTPUT] section, the H-bond list is generated. When using the control file above, the ’hb_listfile‘ will be as follows:
# snapshot | info for analysis atom .. info for target_atom | HB_dis DHA_ang HDA_ang
1 | O PHE 4 BPTI .. OH2 TIP3 2065 WT1 | 2.629 167.111 8.231
1 | OE2 GLU 7 BPTI .. OH2 TIP3 2065 WT1 | 2.796 160.046 13.244
1 | N ASN 43 BPTI .. OH2 TIP3 2065 WT1 | 3.119 140.923 27.451
2 | O PHE 4 BPTI .. OH2 TIP3 2065 WT1 | 2.877 162.791 11.560
2 | OE2 GLU 7 BPTI .. OH2 TIP3 2065 WT1 | 3.099 154.963 17.526
2 | N ASN 43 BPTI .. OH2 TIP3 2065 WT1 | 3.141 138.608 29.278
......
In hb_listfiles, snapshot numbers are written in the leftmost column. In the 2nd-9th columns, atom name, residue name, residue number and segment name are respectively written both for the ‘analysis atom’ and the ‘target atom’. The 1st – 3rd columns from the right in the list represent the geometry of the H-bonds: H-bond distance, DHA_angle and HDA_angle.
case 2: Examine the frequency of H-bond formation during the trajectory
When output_type = Count_atom, the frequency of H-bond formation between ‘analysis_atom‘ and ‘target_atom‘ groups is output for each H-bond pair.
[INPUT]
psffile = BPTI_ionize.psf
reffile = BPTI_ionize.pdb
[OUTPUT]
txtfile = BPTI-external_water_H-bonds.txt
[TRAJECTORY]
trjfile1 = run.dcd
md_step1 = 10
mdout_period1 = 1
ana_period1 = 1
trj_format = DCD
trj_type = COOR+BOX
[SELECTION]
group1 = sid:BPTI & resno:35
group2 = resname:TIP3
[OPTION]
check_only = NO
output_type = Count_atom # number of H-bonds is output per each H-bond pair
analysis_atom = 1
target_atom = 2
solvent_list = TIP3 DPPC # residue names of solvent molecules
# (Here, DPPC is added to show how to list more than one solvent type)
boundary_type = PBC
hb_distance = 3.4
dha_angle = 120.0
hda_angle = 30.0
The option ‘solvent_list‘ is used not to distinguish individual solvent molecules (i.e. water or lipid molecules) in examining the frequency of the H-bond formation between protein and solvent molecules. For example, when you use the control file above, the output file will be as follow:
15 | OH TYR 35 BPTI .. OH2 TIP3
In the output files, the leftmost column represents the total number of the H-bonds between BPTI Tyr35 OH atoms and water molecules during the 10 snapshots used in the analysis.
On the other hand, when ‘solvent_list‘ is blank or not specified, the output file will be as follows:
1 | OH TYR 35 BPTI .. OH2 TIP3 863 WT1
1 | OH TYR 35 BPTI .. OH2 TIP3 1932 WT1
1 | OH TYR 35 BPTI .. OH2 TIP3 2224 WT1
2 | OH TYR 35 BPTI .. OH2 TIP3 2648 WT1
1 | OH TYR 35 BPTI .. OH2 TIP3 3988 WT1
1 | OH TYR 35 BPTI .. OH2 TIP3 4589 WT1
1 | OH TYR 35 BPTI .. OH2 TIP3 5122 WT1
1 | OH TYR 35 BPTI .. OH2 TIP3 6152 WT1
1 | OH TYR 35 BPTI .. OH2 TIP3 7402 WT1
1 | OH TYR 35 BPTI .. OH2 TIP3 8250 WT1
1 | OH TYR 35 BPTI .. OH2 TIP3 9245 WT1
1 | OH TYR 35 BPTI .. OH2 TIP3 4244 WT2
1 | OH TYR 35 BPTI .. OH2 TIP3 4476 WT3
1 | OH TYR 35 BPTI .. OH2 TIP3 8023 WT7
In the output files, the leftmost column represents the total number of the H-bonds between BPTI Tyr35 OH atoms and each water molecule during the 10 snapshots used in the analysis.
